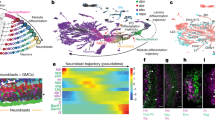
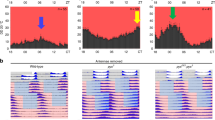
Abstract
Behavioral circadian rhythms are controlled by a neuronal circuit consisting of diverse neuronal subgroups. To understand the molecular mechanisms underlying the roles of neuronal subgroups within the Drosophila circadian circuit, we used cell-type specific gene-expression profiling and identified a large number of genes specifically expressed in all clock neurons or in two important subgroups. Moreover, we identified and characterized two circadian genes, which are expressed specifically in subsets of clock cells and affect different aspects of rhythms. The transcription factor Fer2 is expressed in ventral lateral neurons; it is required for the specification of lateral neurons and therefore their ability to drive locomotor rhythms. The Drosophila melanogaster homolog of the vertebrate circadian gene nocturnin is expressed in a subset of dorsal neurons and mediates the circadian light response. The approach should also enable the molecular dissection of many different Drosophila neuronal circuits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Kaneko, M., Helfrich-Forster, C. & Hall, J.C. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemakers candidates and novel features of clock gene product cycling. J. Neurosci. 17, 6745–6760 (1997).
Mazzoni, E.O., Desplan, C. & Blau, J. Circadian pacemaker neurons transmit and modulate visual information to control a rapid behavioral response. Neuron 45, 293–300 (2005).
Grima, B., Chelot, E., Xia, R. & Rouyer, F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873 (2004).
Stoleru, D., Peng, Y., Agosto, J. & Rosbash, M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868 (2004).
Yu, W. & Hardin, P.E. Circadian oscillators of Drosophila and mammals. J. Cell Sci. 119, 4793–4795 (2006).
Gallego, M. & Virshup, D.M. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148 (2007).
Ceriani, M.F. et al. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22, 9305–9319 (2002).
Claridge-Chang, A. et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32, 657–671 (2001).
Keegan, K.P., Pradhan, S., Wang, J.P. & Allada, R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput. Biol. 3, e208 (2007).
Lin, Y. et al. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99, 9562–9567 (2002).
McDonald, M.J. & Rosbash, M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107, 567–578 (2001).
Ueda, H.R. et al. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 277, 14048–14052 (2002).
Park, J.H. et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. USA 97, 3608–3613 (2000).
Rutila, J.E. et al. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814 (1998).
Kloss, B. et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell 94, 97–107 (1998).
Hempel, C.M., Sugino, K. & Nelson, S.B. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat. Protoc. 2, 2924–2929 (2007).
Sugino, K. et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 9, 99–107 (2006).
Kadener, S., Stoleru, D., McDonald, M., Nawathean, P. & Rosbash, M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 21, 1675–1686 (2007).
Murad, A., Emery-Le, M. & Emery, P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron 53, 689–701 (2007).
Stoleru, D. et al. The Drosophila circadian network is a seasonal timer. Cell 129, 207–219 (2007).
Moore, A.W., Barbel, S., Jan, L.Y. & Jan, Y.N. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc. Natl. Acad. Sci. USA 97, 10436–10441 (2000).
Krapp, A. et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 12, 3752–3763 (1998).
Hoshino, M. et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47, 201–213 (2005).
Green, C.B. & Besharse, J.C. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc. Natl. Acad. Sci. USA 93, 14884–14888 (1996).
Wang, Y. et al. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev. Biol. 1, 9 (2001).
Garbarino-Pico, E. et al. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA 13, 745–755 (2007).
Green, C.B. et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc. Natl. Acad. Sci. USA 104, 9888–9893 (2007).
Konopka, R.J., Pittendrigh, C. & Orr, D. Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. J. Neurogenet. 6, 1–10 (1989).
Roy, P.J., Stuart, J.M., Lund, J. & Kim, S.K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418, 975–979 (2002).
Doyle, J.P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Yang, Z., Edenberg, H.J. & Davis, R.L. Isolation of mRNA from specific tissues of Drosophila by mRNA tagging. Nucleic Acids Res. 33, e148 (2005).
Hoopfer, E.D., Penton, A., Watts, R.J. & Luo, L. Genomic analysis of Drosophila neuronal remodeling: a role for the RNA-binding protein Boule as a negative regulator of axon pruning. J. Neurosci. 28, 6092–6103 (2008).
Finkel, T. Oxygen radicals and signaling. Curr. Opin. Cell Biol. 10, 248–253 (1998).
Lin, F.J., Song, W., Meyer-Bernstein, E., Naidoo, N. & Sehgal, A. Photic signaling by cryptochrome in the Drosophila circadian system. Mol. Cell. Biol. 21, 7287–7294 (2001).
Sathyanarayanan, S. et al. Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen. Genes Dev. 22, 1522–1533 (2008).
Rieger, D., Shafer, O.T., Tomioka, K. & Helfrich-Forster, C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 26, 2531–2543 (2006).
Picot, M., Cusumano, P., Klarsfeld, A., Ueda, R. & Rouyer, F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 5, e315 (2007).
Bahn, J.H., Lee, G. & Park, J.H. Comparative analysis of Pdf-mediated circadian behaviors between Drosophila melanogaster and D. virilis. Genetics 181, 965–975 (2009).
Gronke, S., Bickmeyer, I., Wunderlich, R., Jackle, H. & Kuhnlein, R.P. curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin. Genetics 183, 219–232 (2009).
Emery, P. et al. Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26, 493–504 (2000).
Yoshii, T., Todo, T., Wulbeck, C., Stanewsky, R. & Helfrich-Forster, C. Cryptochrome is present in the compound eyes and a subset of Drosophila's clock neurons. J. Comp. Neurol. 508, 952–966 (2008).
Jiang, S.A., Campusano, J.M., Su, H. & O'Dowd, D.K. Drosophila mushroom body Kenyon cells generate spontaneous calcium transients mediated by PLTX-sensitive calcium channels. J. Neurophysiol. 94, 491–500 (2005).
Kuppers-Munther, B. et al. A new culturing strategy optimises Drosophila primary cell cultures for structural and functional analyses. Dev. Biol. 269, 459–478 (2004).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Kaneko, M. & Hall, J.C. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422, 66–94 (2000).
Klarsfeld, A. et al. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J. Neurosci. 24, 1468–1477 (2004).
Kishiro, Y., Kagawa, M., Naito, I. & Sado, Y. A novel method of preparing rat-monoclonal antibody-producing hybridomas by using rat medial iliac lymph node cells. Cell Struct. Funct. 20, 151–156 (1995).
Shafer, O.T., Rosbash, M. & Truman, J.W. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 22, 5946–5954 (2002).
Houl, J.H., Ng, F., Taylor, P. & Hardin, P.E. CLOCK expression identifies developing circadian oscillator neurons in the brains of Drosophila embryos. BMC Neurosci. 9, 119 (2008).
Acknowledgements
We thank P. Hardin (Texas A&M University) for antisera to CLK as well as the Bloomington Stock Center, National Institute of Genetics of Japan (NIG) and Vienna Drosophila RNAi Center (VDRC) for fly stocks. We are grateful to J. Menet, W. Luo, K. Abruzzi, S. Bradley, L. Griffith, P. Garrity, S. Waddell, R. Allada, Y. Shang, P. Emery and J. Blau for comments on the manuscript. Y. Shang also provided the protocol for culture media preparation, and N. Francis helped with brain dissections. Some of this work was supported by a fellowship to E.N. from the Charles A. King Trust.
Author information
Authors and Affiliations
Contributions
E.N. and M.R. conceived the idea of this project and wrote the manuscript; E.N. conducted molecular and behavioral experiments; K.S. conducted the microarray data analysis; E.K. performed the expression profiling of the adult neurons; K.S. and S.N. contributed to the development of the protocol for the Drosophila cell type–specific expression profiling; E.O. and T.T. generated a NOC-RD specific antibody.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–4 and Supplementary Figures 1–4 (PDF 4836 kb)
Rights and permissions
About this article
Cite this article
Nagoshi, E., Sugino, K., Kula, E. et al. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci 13, 60–68 (2010). https://doi.org/10.1038/nn.2451
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2451
This article is cited by
-
Dopaminergic systems create reward seeking despite adverse consequences
Nature (2023)
-
Maintenance of mitochondrial integrity in midbrain dopaminergic neurons governed by a conserved developmental transcription factor
Nature Communications (2022)
-
Neurofibromin 1 in mushroom body neurons mediates circadian wake drive through activating cAMP–PKA signaling
Nature Communications (2021)
-
A Homeobox Transcription Factor Scarecrow (SCRO) Negatively Regulates Pdf Neuropeptide Expression through Binding an Identified cis-Acting Element in Drosophila melanogaster
Molecular Neurobiology (2020)
-
Enabling cell-type-specific behavioral epigenetics in Drosophila: a modified high-yield INTACT method reveals the impact of social environment on the epigenetic landscape in dopaminergic neurons
BMC Biology (2019)