Abstract
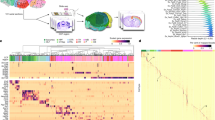
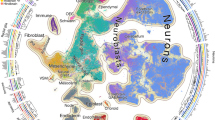
Studying gene expression provides a powerful means of understanding structure-function relationships in the nervous system. The availability of genome-scale in situ hybridization datasets enables new possibilities for understanding brain organization based on gene expression patterns. The Anatomic Gene Expression Atlas (AGEA) is a new relational atlas revealing the genetic architecture of the adult C57Bl/6J mouse brain based on spatial correlations across expression data for thousands of genes in the Allen Brain Atlas (ABA). The AGEA includes three discovery tools for examining neuroanatomical relationships and boundaries: (1) three-dimensional expression-based correlation maps, (2) a hierarchical transcriptome-based parcellation of the brain and (3) a facility to retrieve from the ABA specific genes showing enriched expression in local correlated domains. The utility of this atlas is illustrated by analysis of genetic organization in the thalamus, striatum and cerebral cortex. The AGEA is a publicly accessible online computational tool integrated with the ABA (http://mouse.brain-map.org/agea).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
MacKenzie-Graham, A. et al. The informatics of a C57BL/6J mouse brain atlas. Neuroinformatics 1, 397–410 (2003).
Toga, A.W. & Thompson, P.M. in Brain Mapping: The Systems (eds. Toga, A.W. & Mazziotta, J.C.): 4–5 (Academic Press, San Diego, 2000).
Swanson, L.W. Structure of the Rat Brain (Academic Press, San Diego, 2004).
Paxinos, G. & Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates (Academic Press, San Diego, 2004).
Dong, H.W. The Allen Reference Atlas: A Digital Color Brain Atlas of the C57BL/6J Male Mouse (John Wiley & Sons, Hoboken, New Jersey, USA, 2008).
Ma, Y. et al. A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience 135, 1203–1215 (2005).
Toga, A.W., Thompson, P.M., Mori, S., Amunts, K. & Zilles, K. Towards multimodal atlases of the human brain. Nat. Rev. Neurosci. 7, 952–966 (2006).
MacKenzie-Graham, A. et al. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J. Anat. 204, 93–102 (2004).
Van Essen, D.C., Drury, H.A., Joshi, S. & Miller, M.I. Functional and structural mapping of human cerebral cortex: solutions are in the surfaces. Proc. Natl. Acad. Sci. USA 95, 788–795 (1998).
Gee, C.E. & Roberts, J.L. In situ hybridization histochemistry: a technique for the study of gene expression in single cells. DNA 2, 157–163 (1983).
Visel, A., Thaller, C. & Eichele, G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556 (2004).
Lein, E.S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Sunkin, S.M. & Hohmann, J. G. Insights from spatially mapped gene expression in the mouse brain. Hum. Mol. Genet. 16 (Spec. No. 2): R209–R219 (2007).
Bonner, R.F. et al. Laser capture microdissection: molecular analysis of tissue. Science 278, 1481–1483 (1997).
Zapala, M.A. et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc. Natl. Acad. Sci. USA 102, 10357–10362 (2005).
Flames, N. et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 27, 9682–9695 (2007).
Ng, L.L. et al. Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE Trans. Comput. Biol. Bioinformatics 4, 382–393 (2007).
Price, J.L. in The Rat Nervous System (ed. Paxinos, G.) 632–633 (Academic Press, Sydney, 1995).
Sherman, S.M. & Guillery, R.W. Exploring the Thalamus and its Role in Cortical Function (MIT Press, Cambridge, Massachusetts, USA, 2006).
Nakamura, Y., Otake, K. & Tokuno, H. The parafascicular nucleus relays spinal inputs to the striatum: an electron microscope study in the rat. Neurosci. Res. 56, 73–79 (2006).
Marini, G., Pianca, L. & Tredici, G. Thalamocortical projection from the parafascicular nucleus to layer V pyramidal cells in frontal and cingulate areas of the rat. Neurosci. Lett. 203, 81–84 (1996).
Voorn, P., Vanderschuren, L.J., Groenewegen, H.J., Robbins, T.W. & Pennartz, C.M. Putting a spin on the dorsal-ventral divide of the striatum. Trends Neurosci. 27, 468–474 (2004).
Brodmann, K. in Some Papers on the Cerebral Cortex [originally published 1909; translated as On the comparative localization of the cortex] 201–230 (Thomas, Springfield, Illinois, USA, 1960).
Yamamori, T. & Rockland, K.S. Neocortical areas, layers, connections, and gene expression. Neurosci. Res. 55, 11–27 (2006).
Mountcastle, V.B. Perceptual Neuroscience: The Cerebral Cortex (Harvard University Press, Cambridge, Massachusetts, USA, 1998).
Kruskal, J.B. Non-metric multidimensional scaling: a numerical method. Psychometrika 29, 115–129 (1964).
Paxinos, G. The Rat Nervous System (Academic Press, New York, 1995).
Alvarez-Buylla, A. & Garcia-Verdugo, J.M. Neurogenesis in adult subventricular zone. J. Neurosci. 22, 629–634 (2002).
Abrous, D.N., Koehl, M. & Le Moal, M. Adult neurogenesis: from precursors to network and physiology. Physiol. Rev. 85, 523–569 (2005).
Yushkevich, P.A. et al. Using MRI to build a 3D reference atlas of the mouse brain from histological images. Proc. Intl. Soc. Magn. Reson. Med. Annu. Mtg. 13, 2809 (2005).
Kruskal, J.B. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 29, 1–27 (1964).
DeGroot, M.H. & Schervish, M.J. Probability and Statistics (Addison-Wesley, Boston, 2002).
Acknowledgements
This work was sponsored by the Allen Institute for Brain Science. The authors wish to thank the Allen Institute founders, P.G. Allen and J. Patton, for their vision, encouragement and support. The authors also thank D. Haynor of the University of Washington, Department of Neuroradiology, and C. Thompson of the Allen Institute.
Author information
Authors and Affiliations
Contributions
L.N., CL. and C.D. built the AGEA application; LN., A.B, J.W.B., H.B., P.P.M. and M.H. performed the analyses; H.-W.D., L.P. and J.H. interpreted neuroanatomy; L.K. and S.P. provided informatics support; E.S.L., D.J.A., A.J.R., M.H. provided overall guidance; and L.N., C.C.O., A.B., S.M.S. and M.H. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Methods (including Supplementary Figures 1–4) and Supplementary Results (including Supplementary Figures 5–10 and Supplementary Tables 1–7) (PDF 5131 kb)
Rights and permissions
About this article
Cite this article
Ng, L., Bernard, A., Lau, C. et al. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci 12, 356–362 (2009). https://doi.org/10.1038/nn.2281
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2281
This article is cited by
-
Gold nanoparticle-enhanced X-ray microtomography of the rodent reveals region-specific cerebrospinal fluid circulation in the brain
Nature Communications (2023)
-
DeepSlice: rapid fully automatic registration of mouse brain imaging to a volumetric atlas
Nature Communications (2023)
-
Neurofilament light chain plasma levels are associated with area of brain damage in experimental cerebral malaria
Scientific Reports (2022)
-
Functional orderly topography of brain networks associated with gene expression heterogeneity
Communications Biology (2022)
-
Overcoming false-positive gene-category enrichment in the analysis of spatially resolved transcriptomic brain atlas data
Nature Communications (2021)