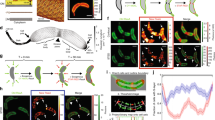
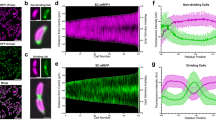
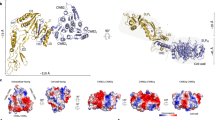
Abstract
Many prokaryotic cells are encapsulated by a surface layer (S-layer) consisting of repeating units of S-layer proteins. S-layer proteins are a diverse class of molecules found in Gram-positive and Gram-negative bacteria and most archaea1–5. S-layers protect cells from the outside, provide mechanical stability and also play roles in pathogenicity. In situ structural information about this highly abundant class of proteins is scarce, so atomic details of how S-layers are arranged on the surface of cells have remained elusive. Here, using purified Caulobacter crescentus' sole S-layer protein RsaA, we obtained a 2.7 Å X-ray structure that shows the hexameric S-layer lattice. We also solved a 7.4 Å structure of the S-layer through electron cryotomography and sub-tomogram averaging of cell stalks. The X-ray structure was docked unambiguously into the electron cryotomography map, resulting in a pseudo-atomic-level description of the in vivo S-layer, which agrees completely with the atomic X-ray lattice model. The cellular S-layer atomic structure shows that the S-layer is porous, with a largest gap dimension of 27 Å, and is stabilized by multiple Ca2+ ions bound near the interfaces. This study spans different spatial scales from atoms to cells by combining X-ray crystallography with electron cryotomography and sub-nanometre-resolution sub-tomogram averaging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Albers, S. V. & Meyer, B. H. The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426 (2011).
Fagan, R. P. & Fairweather, N. F. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222 (2014).
Glauert, A. M. The fine structure of bacteria. Br. Med. Bull. 18, 245–250 (1962).
Sara, M. & Sleytr, U. B. S-layer proteins. J. Bacteriol. 182, 859–868 (2000).
Zhu, C. et al. Diversity in S-layers. Prog. Biophys. Mol. Biol. 123, 1–15 (2017).
Kirk, J. A., Banerji, O. & Fagan, R. P. Characteristics of the Clostridium difficile cell envelope and its importance in therapeutics. Microb. Biotechnol. 10, 76–90 (2016).
Sleytr, U. B. & Beveridge, T. J. Bacterial S-layers. Trends Microbiol. 7, 253–260 (1999).
Houwink, A. L. A macromolecular mono-layer in the cell wall of Spirillum spec. Biochim. Biophys. Acta 10, 360–366 (1953).
Sleytr, U. B. & Glauert, A. M. Ultrastructure of the cell walls of two closely related Clostridia that possess different regular arrays of surface subunits. J. Bacteriol. 126, 869–882 (1976).
Baumeister, W., Wildhaber, I. & Phipps, B. M. Principles of organization in eubacterial and archaebacterial surface proteins. Can. J. Microbiol. 35, 215–227 (1989).
Kessel, M., Wildhaber, I., Cohen, S. & Baumeister, W. Three-dimensional structure of the regular surface glycoprotein layer of Halobacterium volcanii from the Dead Sea. EMBO J. 7, 1549–1554 (1988).
Lupas, A. et al. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176, 1224–1233 (1994).
Baranova, E. et al. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature 487, 119–122 (2012).
Arbing, M. A. et al. Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans. Proc. Natl Acad. Sci. USA 109, 11812–11817 (2012).
Jing, H. et al. Archaeal surface layer proteins contain beta propeller, PKD, and beta helix domains and are related to metazoan cell surface proteins. Structure 10, 1453–1464 (2002).
Kern, J. et al. Structure of surface layer homology (SLH) domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286, 26042–26049 (2011).
Jiang, C., Brown, P. J., Ducret, A. & Brun, Y. V. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506, 489–493 (2014).
Wagner, J. K. & Brun, Y. V. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol. Microbiol. 64, 28–33 (2007).
Amat, F. et al. Analysis of the intact surface layer of Caulobacter crescentus by cryo-electron tomography. J. Bacteriol. 192, 5855–5865 (2010).
Smit, J., Engelhardt, H., Volker, S., Smith, S. H. & Baumeister, W. The S-layer of Caulobacter crescentus: three-dimensional image reconstruction and structure analysis by electron microscopy. J. Bacteriol. 174, 6527–6538 (1992).
Ford, M. J., Nomellini, J. F. & Smit, J. S-layer anchoring and localization of an S-layer-associated protease in Caulobacter crescentus. J. Bacteriol. 189, 2226–2237 (2007).
Nomellini, J. F., Kupcu, S., Sleytr, U. B. & Smit, J. Factors controlling in vitro recrystallization of the Caulobacter crescentus paracrystalline S-layer. J. Bacteriol. 179, 6349–6354 (1997).
Garnham, C. P., Campbell, R. L. & Davies, P. L. Anchored clathrate waters bind antifreeze proteins to ice. Proc. Natl Acad. Sci. USA 108, 7363–7367 (2011).
Ireland, M. M., Karty, J. A., Quardokus, E. M., Reilly, J. P. & Brun, Y. V. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol. Microbiol. 45, 1029–1041 (2002).
Bharat, T. A. M. & Scheres, S. H. W. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat. Protoc. 11, 2054–2065 (2016).
Schur, F. K. M. et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353, 506–508 (2016).
Howorka, S. Rationally engineering natural protein assemblies in nanobiotechnology. Curr. Opin. Biotechnol. 22, 485–491 (2011).
Mark, S. S. et al. Bionanofabrication of metallic and semiconductor nanoparticle arrays using S-layer protein lattices with different lateral spacings and geometries. Langmuir 22, 3763–3774 (2006).
Chang, Y.-W. et al. Architecture of the type IVa pilus machine. Science 351, aad2001 (2016).
Poindexter, J. S. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28, 231–295 (1964).
Stock, D., Perisic, O. & Löwe, J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog. Biophys. Mol. Biol. 88, 311–327 (2005).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D 67, 282–292 (2011).
Sheldrick, G. M. in Direct Methods for Solving Macromolecular Structures (ed. Fortier, S. ) 401–411 (Springer, 1998).
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007).
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. D 66, 470–478 (2010).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006).
Turk, D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr. D 69, 1342–1357 (2013).
Adams, P. D. et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Hagen, W. J. H., Wan, W. & Briggs, J. A. G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2016).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Bharat, T. A. M. et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 9, e1001196 (2011).
Bharat, T. A. M., Russo, C. J., Löwe, J., Passmore, L. A. & Scheres, S. H. W. Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure 23, 1743–1753 (2015).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Briggs, J. A. et al. Structure and assembly of immature HIV. Proc. Natl Acad. Sci. USA 106, 11090–11095 (2009).
Förster, F., Medalia, O., Zauberman, N., Baumeister, W. & Fass, D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl Acad. Sci. USA 102, 4729–4734 (2005).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005).
Acknowledgements
The authors thank M. Skehel and F. Begum (MRC Laboratory of Molecular Biology, LMB) for mass-spectrometric identification of proteins, M. Yu (LMB) for help with X-ray data collection, C. Savva (LMB) for help with cryo-EM data collection, S. Scheres (LMB) for help with RELION software, F. Schur and W. Wan (European Molecular Biology Laboratory, EMBL) for providing high-resolution image-processing code and scripts before publication and for advice on their implementation, F. van den Ent (LMB) for advice on protein purification and T. Darling and J. Grimmett (LMB) for help with high-performance computing. The authors also thank Y. Modis (Cambridge University) for pointing out the similarity to anti-freeze proteins. Part of this work was funded by the European Molecular Biology Organization (aALTF 778-2015 to T.A.M.B.), the Medical Research Council (U105184326 to J.L.), the Wellcome Trust (095514/Z/11/Z to J.L.) and the National Institutes of Health (GM51986 to Y.V.B.). This work was supported by iNEXT, project no. 1482, funded by the Horizon 2020 programme of the European Union.
Author information
Authors and Affiliations
Contributions
T.A.M.B., Y.V.B. and J.L. designed the research. T.A.M.B., D.K.-C., G.G.H., E.W.Y., J.M.D., W.J.H.H. and J.L. performed experiments. W.J.H.H. and J.A.G.B. supported high-resolution cryo-ET and image processing. TA.M.B., D.K.-C., J.M.D., E.W.Y. and J.L. analysed the data. T.A.M.B. and J.L. wrote the manuscript with support from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2; Supplementary Figures 1–7; legends for Supplementary Videos (PDF 2780 kb)
Supplementary Video 1
Whole cell cryo-ET of a C. crescentus cell (related to Figure 1). (AVI 21817 kb)
Supplementary Video 2
Cryo-ET of reconstituted 2D sheets of RsaA (related to Figure 2). (AVI 18787 kb)
Supplementary Video 3
The X-ray structure and map of a RsaA monomer (related to Figure 2). (MOV 35083 kb)
Supplementary Video 4
X-ray crystallography structure of the outer S-layer lattice (related to Figure 2). (MOV 35508 kb)
Supplementary Video 5
Cryo-ET of a C. crescentus stalk (related to Figure 3). (AVI 5543 kb)
Supplementary Video 7
The structure and arrangement of the S-layer on cells (related to Figure 4). (AVI 46055 kb)
Rights and permissions
About this article
Cite this article
Bharat, T., Kureisaite-Ciziene, D., Hardy, G. et al. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat Microbiol 2, 17059 (2017). https://doi.org/10.1038/nmicrobiol.2017.59
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2017.59
This article is cited by
-
Cell cycle dependent coordination of surface layer biogenesis in Caulobacter crescentus
Nature Communications (2024)
-
Structure and function of the EA1 surface layer of Bacillus anthracis
Nature Communications (2023)
-
Bacterial defences: mechanisms, evolution and antimicrobial resistance
Nature Reviews Microbiology (2023)
-
Supramolecular assembly of protein building blocks: from folding to function
Nano Convergence (2022)
-
Structure and activity of particulate methane monooxygenase arrays in methanotrophs
Nature Communications (2022)