Abstract
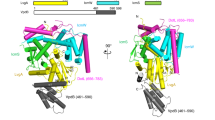
Many bacteria, including Legionella pneumophila, rely on the type IV secretion system to translocate a repertoire of effector proteins into the hosts for their survival and growth. Type IV coupling protein (T4CP) is a hexameric ATPase that links translocating substrates to the transenvelope secretion conduit. Yet, how a large number of effector proteins are selectively recruited and processed by T4CPs remains enigmatic. DotL, the T4CP of L. pneumophila, contains an ATPase domain and a C-terminal extension whose function is unknown. Unlike T4CPs involved in plasmid DNA translocation, DotL appeared to function by forming a multiprotein complex with four other proteins. Here, we show that the C-terminal extension of DotL interacts with DotN, IcmS, IcmW and an additionally identified subunit LvgA, and that this pentameric assembly binds Legionella effector proteins. We determined the crystal structure of this assembly and built an architecture of the T4CP holocomplex by combining a homology model of the ATPase domain of DotL. The holocomplex is a hexamer of a bipartite structure composed of a membrane-proximal ATPase domain and a membrane-distal substrate-recognition assembly. The presented information demonstrates the architecture and functional dissection of the multiprotein T4CP complexes and provides important insights into their substrate recruitment and processing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wallden, K., Rivera-Calzada, A. & Waksman, G. Microreview: type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12, 1203–1212 (2010).
Vincent, C. D., Friedman, J. R., Jeong, K. C., Sutherland, M. C. & Vogel, J. P. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 85, 378–391 (2012).
Gomis-Ruth, F. X., Sola, M., de la Cruz, F. & Coll, M. Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des. 10, 1551–1565 (2004).
Zechner, E. L., Lang, S. & Schildbach, J. F. Assembly and mechanisms of bacterial type IV secretion machines. Phil. Trans. R. Soc. Lond. B 367, 1073–1087 (2012).
Vergunst, A. C. et al. Virb/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982 (2000).
Schrammeijer, B., den Dulk-Ras, A., Vergunst, A. C., Jacome, E. J. & Hooykaas, P. J. J. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 31, 860–868 (2003).
Atmakuri, K., Cascales, E. & Christie, P. J. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54, 1199–1211 (2004).
Jakubowski, S. J., Krishnamoorthy, V., Cascales, E. & Christie, P. J. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341, 961–977 (2004).
Isberg, R. R., O'Connor, T. J. & Heidtman, M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 (2009).
Finsel, I. & Hilbi, H. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell. Microbiol. 17, 935–950 (2015).
Berger, K. H. & Isberg, R. R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19 (1993).
Marra, A., Blander, S. J., Horwitz, M. A. & Shuman, H. A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl Acad. Sci. USA 89, 9607–9611 (1992).
Segal, G., Purcell, M. & Shuman, H. A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl Acad. Sci. USA 95, 1669–1674 (1998).
Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 (1998).
Juhas, M., Crook, D. W. & Hood, D. W. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10, 2377–2386 (2008).
Christie, P. J. & Vogel, J. P. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8, 354–360 (2000).
Vincent, C. D. et al. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 62, 1278–1291 (2006).
Buscher, B. A. et al. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J. Bacteriol. 187, 2927–2938 (2005).
Gomis-Ruth, F. X., Moncalian, G., de la Cruz, F. & Coll, M. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein—detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277, 7556–7566 (2002).
Sutherland, M. C., Nguyen, T. L., Tseng, V. & Vogel, J. P. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog. 8, e1002910 (2012).
Cambronne, E. D. & Roy, C. R. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3, e188 (2007).
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Alva, V., Nam, S. Z., Soding, J. & Lupas, A. N. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 44, W410–W415 (2016).
Edelstein, P. H., Hu, B., Higa, F. & Edelstein, M. A. C. lvgA, a novel Legionella pneumophila virulence factor. Infect. Immun. 71, 2394–2403 (2003).
Vincent, C. D. & Vogel, J. P. The Legionella pneumophila IcmS–LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol. Microbiol. 61, 596–613 (2006).
Fiser, A., Do, R. K. & Sali, A. Modeling of loops in protein structures. Protein Sci. 9, 1753–1773 (2000).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 5.6.1–5.6.37 (2014).
Bardill, J. P., Miller, J. L. & Vogel, J. P. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 56, 90–103 (2005).
Coers, J. et al. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38, 719–736 (2000).
Habyarimana, F., Price, C. T., Santic, M., Al-Khodor, S. & Kwaik, Y. A. Molecular characterization of the Dot/Icm-translocated AnkH and AnkJ eukaryotic-like effectors of Legionella pneumophila. Infect. Immun. 78, 1123–1134 (2010).
Ninio, S., Zuckman-Cholon, D. M., Cambronne, E. D. & Roy, C. R. The Legionella IcmS–IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55, 912–926 (2005).
Jain, A. et al. Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488 (2011).
Ninio, S., Celli, J. & Roy, C. R. A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 5, e1000278 (2009).
Gomis-Rüth, F. X. et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409, 637–641 (2001).
Lu, J. et al. Structural basis of specific TraD–TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. 70, 89–99 (2008).
Atmakuri, K., Ding, Z. Y. & Christie, P. J. Vire2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49, 1699–1713 (2003).
Whitaker, N. et al. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pKM101-encoded conjugation machine. J. Bacteriol. 198, 2701–2718 (2016).
Whitaker, N. et al. The all-alpha domains of coupling proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-encoded type IV secretion systems confer specificity to binding of cognate DNA substrates. J. Bacteriol. 197, 2335–2349 (2015).
Voth, D. E., Broederdorf, L. J. & Graham, J. G. Bacterial type IV secretion systems: versatile virulence machines. Future Microbiol. 7, 241–257 (2012).
Bennett, J. C. Q. & Hughes, C. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 8, 202–204 (2000).
Parsot, C., Hamiaux, C. & Page, A. L. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6, 7–14 (2003).
Stebbins, C. E. & Galan, J. E. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414, 77–81 (2001).
Nagai, H. et al. A C-terminal translocation signal required for Dot/lcm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl Acad. Sci. USA 102, 826–831 (2005).
Huang, L. et al. The E block motif is associated with Legionella pneumophila translocated substrates. Cell. Microbiol. 13, 227–245 (2011).
Lifshitz, Z. et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl Acad. Sci. USA 110, E707–E715 (2013).
Sexton, J. A., Yeo, H. J. & Vogel, J. P. Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol. Microbiol. 57, 70–84 (2005).
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001).
Shen, A. et al. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat. Chem. Biol. 5, 469–478 (2009).
Adams, P. D. et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Mccoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 (1997).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Svergun, D. I., Petoukhov, M. V. & Koch, M. H. J. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 (2001).
Svergun, D., Barberato, C. & Koch, M. H. J. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995).
Semenyuk, A. V. & Svergun, D. I. GNOM—a program package for small-angle scattering data-processing. J. Appl. Crystallogr. 24, 537–540 (1991).
Lee, N. K. et al. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 88, 2939–2953 (2005).
Kim, C., Lee, O. C., Kim, J. Y., Sung, W. & Lee, N. K. Dynamic release of bending stress in short dsDNA by formation of a kink and forks. Angew. Chem. Int. Ed. 54, 8943–8947 (2015).
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Jain, A., Liu, R., Xiang, Y. K. & Ha, T. Single-molecule pull-down for studying protein interactions. Nat. Protoc. 7, 445–452 (2012).
Joo, K. et al. Template based protein structure modeling by global optimization in CASP11. Proteins 84(Suppl. 1), 221–232 (2016).
Joo, K., Lee, J., Kim, I., Lee, S. J. & Lee, J. Multiple sequence alignment by conformational space annealing. Biophys. J. 95, 4813–4819 (2008).
Lee, J., Scheraga, H. A. & Rackovsky, S. New optimization method for conformational energy calculations on polypeptides: conformational space annealing. J. Comput. Chem. 18, 1222–1232 (1997).
Lee, J., Lee, I. H. & Lee, J. Unbiased global optimization of Lennard–Jones clusters for N < or = 201 using the conformational space annealing method. Phys. Rev. Lett. 91, 080201 (2003).
Joo, K. et al. All-atom chain-building by optimizing MODELLER energy function using conformational space annealing. Proteins 75, 1010–1023 (2009).
Krivov, G. G., Shapovalov, M. V. & Dunbrack, R. L. Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 (2009).
Acknowledgements
This study made use of Beamlines 4C and 5C at Pohang Accelerator Laboratory, Korea, and was supported by the National Research Foundation of Korea (NRF) (grant no. 2008-00576) and KAIST Future Systems Healthcare Project, Ministry of Science, ICT and Future Planning. J.D.K. was supported by a TJ PARK postdoctoral fellowship from POSCO TJ PARK Foundation. The authors thank the Korea Institute for Advanced Study for providing computing resources (KIAS Center for Advanced Computation Linux Cluster).
Author information
Authors and Affiliations
Contributions
M.-J.K., J.D.K., H.K., S.K. and Y.-G.K. performed X-ray crystallography and biochemical experiments. C.K. conducted the ALEX-FRET experiment, J.W.B. the SiMPull, K.J. and J.L. homology modelling, and K.S.J. the SAXS. B.-H.O., M.-J.K., J.D.K., N.K.L. and J.U.J. conceived the experiments and wrote the manuscript. All authors discussed the results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–7, Supplementary Table 1, Supplementary References. (PDF 7746 kb)
Rights and permissions
About this article
Cite this article
Kwak, MJ., Kim, J., Kim, H. et al. Architecture of the type IV coupling protein complex of Legionella pneumophila. Nat Microbiol 2, 17114 (2017). https://doi.org/10.1038/nmicrobiol.2017.114
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2017.114
This article is cited by
-
Structural and functional diversity of type IV secretion systems
Nature Reviews Microbiology (2024)
-
Genome wide analysis revealed conserved domains involved in the effector discrimination of bacterial type VI secretion system
Communications Biology (2023)
-
Structural basis for effector protein recognition by the Dot/Icm Type IVB coupling protein complex
Nature Communications (2020)
-
Mechanism of effector capture and delivery by the type IV secretion system from Legionella pneumophila
Nature Communications (2020)
-
Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS
Nature Microbiology (2019)