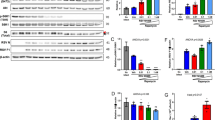
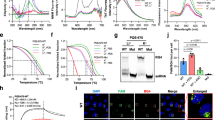
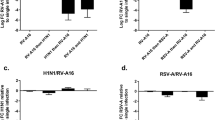
Abstract
Human respiratory syncytial virus (hRSV) is a major cause of morbidity and mortality in the paediatric, elderly and immune-compromised populations1,2. A gap in our understanding of hRSV disease pathology is the interplay between virally encoded immune antagonists and host components that limit hRSV replication. hRSV encodes for non-structural (NS) proteins that are important immune antagonists3–6; however, the role of these proteins in viral pathogenesis is incompletely understood. Here, we report the crystal structure of hRSV NS1 protein, which suggests that NS1 is a structural paralogue of hRSV matrix (M) protein. Comparative analysis of the shared structural fold with M revealed regions unique to NS1. Studies on NS1 wild type or mutant alone or in recombinant RSVs demonstrate that structural regions unique to NS1 contribute to modulation of host responses, including inhibition of type I interferon responses, suppression of dendritic cell maturation and promotion of inflammatory responses. Transcriptional profiles of A549 cells infected with recombinant RSVs show significant differences in multiple host pathways, suggesting that NS1 may have a greater role in regulating host responses than previously appreciated. These results provide a framework to target NS1 for therapeutic development to limit hRSV-associated morbidity and mortality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hall, C. B. The burgeoning burden of respiratory syncytial virus among children. Infect. Disord. Drug Targets 12, 92–97 (2012).
Stockman, L. J., Curns, A. T., Anderson, L. J. & Fischer-Langley, G. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997–2006. Pediatr. Infect. Dis. J. 31, 5–9 (2012).
Bitko, V. et al. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-κB-dependent, interferon-independent mechanism and facilitate virus growth. J. Virol. 81, 1786–1795 (2007).
Lo, M. S., Brazas, R. M. & Holtzman, M. J. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79, 9315–9319 (2005).
Spann, K. M., Tran, K. C., Chi, B., Rabin, R. L. & Collins, P. L. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78, 4363–4369 (2004).
Spann, K. M., Tran, K. C. & Collins, P. L. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79, 5353–5362 (2005).
Munoz, F. M. Respiratory syncytial virus in infants: is maternal vaccination a realistic strategy? Curr. Opin. Infect. Dis. 28, 221–224 (2015).
Collins, P. L. & Melero, J. A. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 162, 80–99 (2011).
Simoes, E. A. et al. Challenges and opportunities in developing respiratory syncytial virus therapeutics. J. Infect. Dis. 211 (Suppl. 1), S1–S20 (2015).
Bossert, B., Marozin, S. & Conzelmann, K. K. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77, 8661–8668 (2003).
Ren, J. et al. A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein. J. Gen. Virol. 92, 2153–2159 (2011).
Goswami, R. et al. Viral degradasome hijacks mitochondria to suppress innate immunity. Cell Res. 23, 1025–1042 (2013).
Elliott, J. et al. Respiratory syncytial virus NS1 protein degrades STAT2 by using the elongin-cullin E3 ligase. J. Virol. 81, 3428–3436 (2007).
Straub, C. P. et al. Mutation of the elongin C binding domain of human respiratory syncytial virus non-structural protein 1 (NS1) results in degradation of NS1 and attenuation of the virus. Virol. J. 8, 252 (2011).
Munir, S. et al. Respiratory syncytial virus interferon antagonist NS1 protein suppresses and skews the human T lymphocyte response. PLoS Pathog. 7, e1001336 (2011).
Munir, S. et al. Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J. Virol. 82, 8780–8796 (2008).
Le Nouen, C. et al. Infection and maturation of monocyte-derived human dendritic cells by human respiratory syncytial virus, human metapneumovirus, and human parainfluenza virus type 3. Virology 385, 169–182 (2009).
Becker, Y. Respiratory syncytial virus(RSV)-induced allergy may be controlled by IL-4 and CX3C fractalkine antagonists and CpG ODN as adjuvant: hypothesis and implications for treatment. Virus Genes 33, 253–264 (2006).
Becker, Y. Respiratory syncytial virus (RSV) evades the human adaptive immune system by skewing the Th1/Th2 cytokine balance toward increased levels of Th2 cytokines and IgE, markers of allergy—a review. Virus Genes 33, 235–252 (2006).
Gonzalez, P. A. et al. Respiratory syncytial virus impairs T cell activation by preventing synapse assembly with dendritic cells. Proc. Natl Acad. Sci. USA 105, 14999–15004 (2008).
Hastie, M. L. et al. The human respiratory syncytial virus nonstructural protein 1 regulates type I and type II interferon pathways. Mol. Cell. Proteomics 11, 108–127 (2012).
Wu, W. et al. The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J. Virol. 86, 7777–7789 (2012).
Money, V. A., McPhee, H. K., Mosely, J. A., Sanderson, J. M. & Yeo, R. P. Surface features of a Mononegavirales matrix protein indicate sites of membrane interaction. Proc. Natl Acad. Sci. USA 106, 4441–4446 (2009).
Dessen, A., Volchkov, V., Dolnik, O., Klenk, H. D. & Weissenhorn, W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 19, 4228–4236 (2000).
Battisti, A. J. et al. Structure and assembly of a paramyxovirus matrix protein. Proc. Natl Acad. Sci. USA 109, 13996–14000 (2012).
Gaudier, M., Gaudin, Y. & Knossow, M. Crystal structure of vesicular stomatitis virus matrix protein. EMBO J. 21, 2886–2892 (2002).
Neumann, P. et al. Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties. Proc. Natl Acad. Sci. USA 106, 3710–3715 (2009).
Forster, A., Maertens, G. N., Farrell, P. J. & Bajorek, M. Dimerization of matrix protein is required for budding of respiratory syncytial virus. J. Virol. 89, 4624–4635 (2015).
Holtzman, M. J., Byers, D. E., Alexander-Brett, J. & Wang, X. The role of airway epithelial cells and innate immune cells in chronic respiratory disease. Nat. Rev. Immunol. 14, 686–698 (2014).
Valarcher, J. F. et al. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77, 8426–8439 (2003).
Dave, K. A. et al. A comprehensive proteomic view of responses of A549 type II alveolar epithelial cells to human respiratory syncytial virus infection. Mol. Cell. Proteomics 13, 3250–3269 (2014).
Martinez, I., Lombardia, L., Garcia-Barreno, B., Dominguez, O. & Melero, J. A. Distinct gene subsets are induced at different time points after human respiratory syncytial virus infection of A549 cells. J. Gen. Virol. 88, 570–581 (2007).
Karron, R. A. et al. A gene deletion that up-regulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci. Transl. Med. 7, 312ra175 (2015).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006).
Sheldrick, G. M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 (2010).
Otwinowski, Z. in CCP4 Study Weekend (eds Wolf, W., Evans, P. R. & Leslie, A. G. W. ) 80–86 (Science and Engineering Research Council, 1991).
Cowtan, K. & Main, P. Miscellaneous algorithms for density modification. Acta Crystallogr. D 54, 487–493 (1998).
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Perrakis, A., Harkiolaki, M., Wilson, K. S. & Lamzin, V. S. ARP/wARP and molecular replacement. Acta Crystallogr. D 57, 1445–1450 (2001).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55, 247–255 (1999).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Davis, I. W. et al. Molprobity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994).
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Laskowski, R. A. Enhancing the functional annotation of PDB structures in PDBsum using key figures extracted from the literature. Bioinformatics 23, 1824–1827 (2007).
Laskowski, R. A. & Swindells, M. B. Ligplot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Bond, C. S. Topdraw: a sketchpad for protein structure topology cartoons. Bioinformatics 19, 311–312 (2003).
Thompson, J. D., Gibson, T. J. & Higgins, D. G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2, 2.3 (2002).
Gouet, P., Robert, X. & Courcelle, E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 (2003).
Hotard, A. L. et al. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology 434, 129–136 (2012).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Lee, B. & Richards, F. M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 (1971).
Vitalis, A. & Pappu, R. V. ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. J. Comput. Chem. 30, 673–699 (2009).
Acknowledgements
The authors thank H. Virgin, M. Diamond, T. Ellenberger and J. Payton, M. Dinauer and E.E.L. Amarasinghe for discussions and J. Huh for technical support. Work in our laboratories is supported, in part, by NIH grants (R01AI107056 (to D.W.L.), R01AI123926 (to G.K.A.), R01AI114654 (to C.F.B.), U191099565 (G.K.A. is the PI of the subaward from a U19 grant for which Ting is the PI), U19AI109945 (to C.F.B.), U19AI109664 (to C.F.B.), U19AI070489 (to M.J.H.), R01AI111605 (to M.J.H.), R01 AI130591 (to M.J.H.), R01AI087798 (to M.L.M.), U19AI095227 (to M.L.M.) and T32-CA09547-37 (D.S.J. is the recipient of a training award from a T32 grant for which Allen is the PI)), the Department of Defense, Defense Threat Reduction Agency grants HDTRA1-16-0033 (to C.F.B.) and HDTRA1-16-0033 (to C.F.B.), the National Science Foundation MCB-1121867 (to R.V.P.) and the Children's Discovery Institute PD-II-2013-272 (to G.K.A.). S.C. is funded in part by an American Heart Association Postdoctoral Fellowship (15POST25140009). We thank members in the Amarasinghe, Leung, Basler, Artyomov and Holtzman laboratories and S. Ginell, N. Duke, R. Alkire, K. Lazarski, M. Ficner-Radford, Y. Kim and A. Joachimiak at Argonne National Laboratory SBC Sector 19. Use of Argonne National Laboratory Structural Biology Center beam lines at the Advanced Photon Source is supported by the US Department of Energy under contract DE-AC02-06CH11357. The content of the information does not necessarily reflect the position or the policy of the federal government and no official endorsement should be inferred.
Author information
Authors and Affiliations
Contributions
G.K.A. and D.W.L. conceived and designed the overall study, with input from the co-authors. S.C., P.L., E.E., E.A., B.C.Y., D.M.B., M.R.E., A.M., P.R., D.S.J., G.K.A. and D.W.L. performed research. M.L.M. provided the wild-type virus. All co-authors analysed the results. R.V.P., M.J.H., M.L.M., M.A., C.F.B., G.K.A. and D.W.L. designed and coordinated studies within each group. C.F.B., G.K.A. and D.W.L. wrote the manuscript with input from all co-authors. All authors analysed the results, and read and approved the manuscript for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–12, Supplementary Tables 1 and 2. (PDF 22332 kb)
Rights and permissions
About this article
Cite this article
Chatterjee, S., Luthra, P., Esaulova, E. et al. Structural basis for human respiratory syncytial virus NS1-mediated modulation of host responses. Nat Microbiol 2, 17101 (2017). https://doi.org/10.1038/nmicrobiol.2017.101
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2017.101
This article is cited by
-
Respiratory syncytial virus infection and novel interventions
Nature Reviews Microbiology (2023)
-
The nonstructural proteins of Pneumoviruses are remarkably distinct in substrate diversity and specificity
Virology Journal (2017)