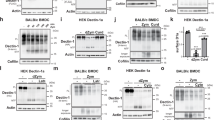
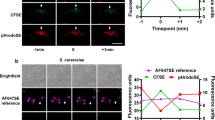
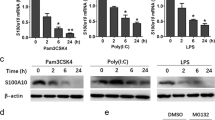
Abstract
Microbial proteases degrade a variety of host proteins1–3. However, it has remained largely unknown why microorganisms have evolved to acquire such proteases and how the host responds to microbially degraded products. Here, we have found that immunoglobulins disrupted by microbial pathogens are specifically detected by leukocyte immunoglobulin-like receptor A2 (LILRA2), an orphan activating receptor expressed on human myeloid cells. Proteases from Mycoplasma hyorhinis, Legionella pneumophila, Streptococcus pneumonia and Candida albicans cleaved the N-terminus of immunoglobulins. Identification of the immunoglobulin-cleaving protease from L. pneumophila revealed that the protease is conserved across some bacteria including Vibrio spp. and Pseudomonas aeruginosa. These microbially cleaved immunoglobulins but not normal immunoglobulins stimulated human neutrophils via LILRA2. In addition, stimulation of primary monocytes via LILRA2 inhibited the growth of L. pneumophila. When mice were infected with L. pneumophila, immunoglobulins were cleaved and recognized by LILRA2. More importantly, cleaved immunoglobulins were detected in patients with bacterial infections and stimulated LILRA2-expressing cells. Our findings demonstrate that LILRA2 is a type of innate immune receptor in the host immune system that detects immunoglobulin abnormalities caused by microbial pathogens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Maeda, H. Role of microbial proteases in pathogenesis. Microbiol. Immunol. 40, 685–699 (1996).
Miyoshi, S. & Shinoda, S. Microbial metalloproteases and pathogenesis. Microbes. Infect. 2, 91–98 (2000).
Finlay, B. B. & McFadden, G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006).
Arase, H. & Lanier, L. L. Specific recognition of virus-infected cells by paired NK receptors. Rev. Med. Virol. 14, 83–93 (2004).
Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B. & Lanier, L. L. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326 (2002).
Satoh, T. et al. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132, 935–944 (2008).
Cosman, D. et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 7, 273–282 (1997).
Smith, H. R. et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl Acad. Sci. USA 99, 8826–8831 (2002).
Kielczewska, A. et al. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J. Exp. Med. 206, 515–523 (2009).
Samaridis, J. & Colonna, M. Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cells: structural evidence for new stimulatory and inhibitory pathways. Eur. J. Immunol. 27, 660–665 (1997).
Nakajima, H., Samaridis, J., Angman, L. & Colonna, M. Human myeloid cells express an activating ILT receptor (ILT1) that associates with Fc receptor γ-chain. J. Immunol. 162, 5–8 (1999).
Brown, D., Trowsdale, J. & Allen, R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens 64, 215–225 (2004).
Hirayasu, K. & Arase, H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J. Hum. Genet. 60, 703–708 (2015).
Colonna, M. et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 186, 1809–1818 (1997).
Zheng, J. et al. Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature 485, 656–660 (2012).
Atwal, J. K. et al. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science 322, 967–970 (2008).
Canavez, F. et al. Comparison of chimpanzee and human leukocyte Ig-like receptor genes reveals framework and rapidly evolving genes. J. Immunol. 167, 5786–5794 (2001).
Hirayasu, K. et al. Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in Northeast Asians. Am. J. Hum. Genet. 82, 1075–1083 (2008).
Bashirova, A. A. et al. Diversity of the human LILRB3/A6 locus encoding a myeloid inhibitory and activating receptor pair. Immunogenetics 66, 1–8 (2014).
Lopez-Alvarez, M. R., Jones, D. C., Jiang, W., Traherne, J. A. & Trowsdale, J. Copy number and nucleotide variation of the LILR family of myelomonocytic cell activating and inhibitory receptors. Immunogenetics 66, 73–83 (2014).
Ohtsuka, M. et al. NFAM1, an immunoreceptor tyrosine-based activation motif-bearing molecule that regulates B cell development and signaling. Proc. Natl Acad. Sci. USA 101, 8126–8131 (2004).
Wang, J. & Arase, H. Regulation of immune responses by neutrophils. Ann. NY Acad. Sci. 1319, 66–81 (2014).
Khweek, A. A. & Amer, A. Replication of Legionella pneumophila in human cells: why are we susceptible? Front. Microbiol. 1, 133 (2010).
Black, W. J., Quinn, F. D. & Tompkins, L. S. Legionella pneumophila zinc metalloprotease is structurally and functionally homologous to Pseudomonas aeruginosa elastase. J. Bacteriol. 172, 2608–2613 (1990).
Takai, T. Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology 115, 433–440 (2005).
Kelley, J., Walter, L. & Trowsdale, J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 1, 129–139 (2005).
Mestas, J. & Hughes, C. C. Of mice and not men: differences between mouse and human immunology. J. Immunol. 172, 2731–2738 (2004).
Willcox, B. E., Thomas, L. M. & Bjorkman, P. J. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nature Immunol. 4, 913–919 (2003).
Allen, R. L., Raine, T., Haude, A., Trowsdale, J. & Wilson, M. J. Leukocyte receptor complex-encoded immunomodulatory receptors show differing specificity for alternative HLA-B27 structures. J. Immunol. 167, 5543–5547 (2001).
Chen, Y. et al. Crystal structure of myeloid cell activating receptor leukocyte Ig-like receptor A2 (LILRA2/ILT1/LIR-7) domain swapped dimer: molecular basis for its non-binding to MHC complexes. J. Mol. Biol. 386, 841–853 (2009).
Brezski, R. J. & Jordan, R. E. Cleavage of IgGs by proteases associated with invasive diseases: an evasion tactic against host immunity? MAbs 2, 212–220 (2010).
Clark, R. A. & Nauseef, W. M. Isolation and functional analysis of neutrophils. Curr. Protoc. Immunol. 7, 7.23 (2001).
Nagai, H. & Roy, C. R. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 20, 5962–5970 (2001).
Homma, M., Chibana, H. & Tanaka, K. Induction of extracellular proteinase in Candida albicans. J. Gen. Microbiol. 139, 1187–1193 (1993).
Shiroishi, M. et al. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J. Biol. Chem. 281, 10439–10447 (2006).
Jiang, Y. et al. Transport of misfolded endoplasmic reticulum proteins to the cell surface by MHC class II molecules. Int. Immunol. 25, 235–246 (2013).
Tiller, T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124 (2008).
Scotto-Lavino, E., Du, G. & Frohman, M. A. 5′ end cDNA amplification using classic RACE. Nature Protoc. 1, 2555–2562 (2006).
Steurer, W. et al. Ex vivo coating of islet cell allografts with murine CTLA4/Fc promotes graft tolerance. J. Immunol. 155, 1165–1174 (1995).
Jin, H. et al. Autoantibodies to IgG/HLA class II complexes are associated with rheumatoid arthritis susceptibility. Proc. Natl Acad. Sci. USA 111, 3787–3792 (2014).
Morita, S., Kojima, T. & Kitamura, T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene. Ther. 7, 1063–1066 (2000).
Moffat, J. F., Black, W. J. & Tompkins, L. S. Further molecular characterization of the cloned Legionella pneumophila zinc metalloprotease. Infect. Immun. 62, 751–753 (1994).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Emmendorffer, A., Hecht, M., Lohmann-Matthes, M. L. & Roesler, J. A fast and easy method to determine the production of reactive oxygen intermediates by human and murine phagocytes using dihydrorhodamine 123. J. Immunol. Methods. 131, 269–275 (1990).
Coers, J., Vance, R. E., Fontana, M. F. & Dietrich, W. F. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 9, 2344–2357 (2007).
Acknowledgements
The authors thank K. Saito of the DNA-chip Development Center for Infectious Diseases (RIMD, Osaka University) for technical assistance, Y. Horiguchi (Department of Molecular Bacteriology, RIMD, Osaka University) for discussions, J. Matsuyama (Pathogenic Microbes Repository Unit, RIMD, Osaka University) for preparing various bacteria and J. Coers (Department of Molecular Genetics and Microbiology and Immunology, Duke University Medical Center) for providing a plasmid for the P. luminescens luxCDABE operon. This work was partially supported by JSPS KAKENHI (grant nos. 15K15131, 15H02545, 24115005, 26117714 and 26870334), the Practical Research Project for Allergic Diseases and Immunology from the Japan Agency for Medical Research and development, AMED, The Mochida Memorial Foundation for Medical and Pharmaceutical Research (to K.H.), The Uehara Memorial Foundation, the Terumo Life Science Foundation and the Tokyo Biochemical Research Foundation (to H.A.).
Author information
Authors and Affiliations
Contributions
K.H. performed experiments, analysed and discussed the data and wrote the manuscript. F.S. assisted with experiments, discussed the data and edited the manuscript. T.S. assisted with experimental design, discussed the data and edited the manuscript. K.S. assisted with experiments and discussed the data. N.A., K.O., T.Y., H.M., I.N., Y.N. and I.K. collected and analysed clinical samples and discussed the data. H.C. assisted with fungal experiments and discussed the data. T.K. and H.N. assisted with bacterial experiments and discussed the data. M.C. assisted with experiments and discussed the data. H.A. designed the study, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-21 and Table 1 (PDF 4728 kb)
Rights and permissions
About this article
Cite this article
Hirayasu, K., Saito, F., Suenaga, T. et al. Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2. Nat Microbiol 1, 16054 (2016). https://doi.org/10.1038/nmicrobiol.2016.54
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.54
This article is cited by
-
Immunoglobulin-like receptors and the generation of innate immune memory
Immunogenetics (2022)
-
Characterization of LILRB3 and LILRA6 allelic variants in the Japanese population
Journal of Human Genetics (2021)
-
Structural basis for RIFIN-mediated activation of LILRB1 in malaria
Nature (2020)
-
Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors
Nature (2017)
-
Sensing broken antibody
Nature Reviews Immunology (2016)