Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) causes sporadic zoonotic disease and healthcare-associated outbreaks in human. MERS is often complicated by acute respiratory distress syndrome (ARDS) and multi-organ failure1,2. The high incidence of renal failure in MERS is a unique clinical feature not often found in other human coronavirus infections3,4. Whether MERS-CoV infects the kidney and how it triggers renal failure are not understood5,6. Here, we demonstrated renal infection and apoptotic induction by MERS-CoV in human ex vivo organ culture and a nonhuman primate model. High-throughput analysis revealed that the cellular genes most significantly perturbed by MERS-CoV have previously been implicated in renal diseases. Furthermore, MERS-CoV induced apoptosis through upregulation of Smad7 and fibroblast growth factor 2 (FGF2) expression in both kidney and lung cells. Conversely, knockdown of Smad7 effectively inhibited MERS-CoV replication and protected cells from virus-induced cytopathic effects. We further demonstrated that hyperexpression of Smad7 or FGF2 induced a strong apoptotic response in kidney cells. Common marmosets infected by MERS-CoV developed ARDS and disseminated infection in kidneys and other organs. Smad7 and FGF2 expression were elevated in the lungs and kidneys of the infected animals. Our results provide insights into the pathogenesis of MERS-CoV and host targets for treatment.
Similar content being viewed by others
The understanding of the pathogenesis of Middle East respiratory syndrome coronavirus (MERS-CoV) leading to pulmonary and renal damage is limited by the lack of postmortem examination in deceased MERS patients. To understand the host response to MERS-CoV infection, we determined the transcriptomic profiles of polarized human bronchial epithelial Calu-3 cells infected with MERS-CoV versus that of severe acute respiratory syndrome coronavirus (SARS-CoV) (Fig. 1a; data available at Sequence Read Archive accession, SRP056612). The gene expression levels of the virus-infected samples at different time points were compared with those of the same samples at time point zero. Our data showed that there were 462, 639, 1,650 and 4,287 differentially expressed genes at 2, 4, 12 and 24 h after MERS-CoV infection, respectively. Remarkably, genes classified under the categories ‘renal necrosis/cell death/nephritis/inflammation/damage/proliferation’ were predominantly affected in MERS-CoV-infected but not SARS-CoV-infected samples. The results correlated with the clinical observation that a high incidence of renal failure was found in MERS but not SARS patients3,7,8.
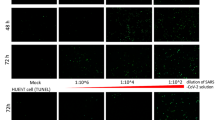
a, Pathway analyses of MERS-CoV-infected (top) and SARS-CoV-infected (bottom) Calu-3 cells based on differential gene expression profiles. Red line: the most significant –log(P value) of the most perturbed biological pathway by SARS-CoV. The –log(P value) is calculated based on the hypergeometric distribution (right-tailed Fisher's exact test) of a gene of interest that can be found in a molecular network over that of a randomly selected gene. Data were derived from two independent experiments. b, Ex vivo human kidney cultures were inoculated with either MERS-CoV or SARS-CoV. The MERS-CoV-inoculated human kidneys were cryosectioned at 24 h post inoculation (top). NP of MERS-CoV was immunostained by specific antibodies (green, left). The kidneys were co-stained with cell-type-specific antibodies against cytokeratin 18 (CK18), CD31 and synaptopodin (red, middle). Co-localization of viral antigens with cellular markers is shown in yellow (right). Nuclei counterstained by DAPI are shown in blue. Representative images from three independent experiments are shown. Total RNAs of MERS-CoV-inoculated (left, lanes 1–5) and SARS-CoV-inoculated (right, lanes 1–5) human kidneys samples were collected at the indicated time points post infection (bottom). Viral RNAs were detected by RT-qPCR as described previously6. MERS-CoV-infected HK2 (left, ‘+’, lane 6) and SARS-CoV-infected Calu-3 samples (right, ‘+’, lane 6) were included as positive controls. Mock-treated HK2 (left, ‘–’, lane 7) and Calu-3 (right, ‘–’, lane 7) samples were included as negative controls. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was also detected as a loading control. c, Conditioned medium (left) and cell lysate (right) of MERS-CoV-infected (M) and SARS-CoV-infected (S) HK2 and NHMC cells were collected 24 h post inoculation and the viruses were quantified using TCID50 assays as described6. C, mock infected; M, MERS-CoV-infected; S, SARS-CoV-infected. Error bars represent the mean ± s.d. of three independent experiments. Samples from a–c represent biological replicates.
Despite reports on the detection of MERS-CoV RNA in the urine of infected patients9,10, in vivo evidence for direct MERS-CoV infection of the kidney is lacking. Hence, we developed an ex vivo human kidney culture model and showed that MERS-CoV, but not SARS-CoV, productively infected kidney cells, as indicated by viral nucleocapsid protein (NP) expression with immunofluorescence (Fig. 1b, top, and Supplementary Fig. 1) and increasing viral load in collected tissues (Fig. 1b, bottom). To specify the infected cell types, we co-stained the kidney tissue for MERS-CoV NP and various kidney cellular markers. The substantial co-localization of MERS-CoV NP with cytokeratin 18 in renal tubular cells, CD31 in vascular epithelial cells and synaptopodin in podocytes suggested that multiple types of kidney cells were susceptible to MERS-CoV infection. The tissue susceptibility was also recapitulated in renal cellular models, including primary normal human mesangial cells (NHMCs) and renal proximal tubular cells (HK2) (Fig. 1c). These models allowed us to further investigate the biological changes induced by MERS-CoV infection in vitro.
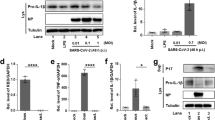
Based on our findings in transcriptomic and ex vivo organ culture studies, we hypothesized that MERS-CoV can induce renal failure through the induction of apoptosis, a major type of cell death. Using a fluorometric assay for apoptotic caspase-3, we detected a strong pro-apoptotic response in MERS-CoV-infected HK2 cells (Fig. 2a). This was confirmed independently by terminal deoxynucleotidyl transferase dUTP nick end labelling assay (Supplementary Fig. 2a). The elevated expression of caspase-3 in MERS-CoV-infected but not mock-infected cells was further confirmed by western blot analysis (Fig. 2b). Because our transcriptomic data indicated that the biological pathways related to ‘renal necrosis/cell death’ were most severely perturbed in MERS-CoV-infected cells (Fig. 1a), we searched for genes that were most prominently affected for further mechanistic studies of MERS-CoV-induced apoptosis. Several criteria were used to identify genes whose expression was specifically modulated by MERS-CoV infection. First, the expression of the affected genes should be progressively and strongly induced throughout the course of MERS-CoV infection. Moreover, their expression should not be restricted only to the lung; they should also be expressed in the kidney. Finally, the expression of the selected genes should be markedly induced by MERS-CoV, but not SARS-CoV, infection (see Methods for details). We selected Smad7 and FGF2, as their expression was most prominently affected when compared with the expression of other deregulated genes not directly related to apoptosis such as clusterin (CLU), superoxide dismutase (SOD1) or catalase (CAT) (Fig. 2c and Supplementary Fig. 3). It should be noted that genes that encode inhibitors of apoptosis, such as baculoviral IAP repeat-containing protein3 (BIRC3), could also play a regulatory role in MERS-CoV biogenesis (Supplementary Fig. 3b,c). Intuitively, the anti-apoptotic activities of BIRC3 may play contributory roles in the early stage of MERS-CoV replication. Moreover, a recent study demonstrated that BIRC3 is required for efficient propagation of the baculovirus Bombyx mori nucleopolyhedrovirus through an unknown mechanism11. Further study will be required to dissect the function(s) of BIRC3 in MERS-CoV replication. Previous reports suggested that Smad7 is significantly associated with renal disease pathogenesis by inducing apoptosis12,13 in mesangial cells and podocytes through caspase 3-dependent14 and -independent pathways15, respectively. More recently, the apoptotic role of Smad7 has been further demonstrated in hepatocarcinogenesis through the attenuation of NFκB and TGFβ signalling16. Similarly, FGF2 induces podocyte injury and glomerulosclerosis in mouse and rat models17,18. The roles of Smad7 and FGF2 in MERS-CoV-induced apoptosis not only in kidneys but also in lungs were supported by their elevated expression in both MERS-CoV-infected HK2 (Fig. 2b,d,e) and Calu-3 cells (Supplementary Fig. 3a). Furthermore, we demonstrated that overexpression of Smad7 and FGF2 in uninfected kidney cells induced apoptosis in a dose-dependent manner (Supplementary Fig. 2b).
a, Apoptosis of MERS-CoV-infected (M) and SARS-CoV-infected (S) HK2 cells were measured by fluorometric analysis of caspase-3 activity. Mock-infected cells (C) were included as negative control. a.u., arbitrary units. b, Western blot analysis of caspase-3, Smad7, FGF2, MERS-CoV NP and SARS-CoV NP in MERS-CoV-, SARS- and mock-infected HK2 cells. γ-Tubulin was also detected as a loading control. Representative images from three independent experiments are shown. c, Heat map showing the candidate genes in the category of ‘renal necrosis/cell death’ identified in Fig. 1a. Arrows point to SMAD7 and FGF2, which were selected for further study. d, The relative expression of Smad7 (left) and FGF2 (right) mRNA was measured using RT-qPCR. e, Quantitative measurement of secreted FGF2 was performed using an anti-FGF2 enzyme-linked immunosorbent assay kit. Statistical significance was evaluated by Student's t-tests and P values are shown in a,d,e. Except for c, all samples were harvested at 24 h.p.i. for measurements. Error bars in a,d,e represent the mean ± s.d. of three independent experiments. Samples from a–e represent biological replicates.
If Smad7 and FGF2 are critical mediators for MERS-CoV pathogenesis, compromising their expression might subvert virus-induced damage. We first knocked down Smad7 mRNA in HK2 cells using siRNAs (Smad7-1 siRNA and Smad7-2 siRNA), which reduced the expression of Smad7 by about 75% at protein levels (Fig. 3a, left top) and about 40% at RNA levels (Fig. 3a, right top). The effect of Smad7 expression on MERS-CoV-induced apoptosis was further determined by measuring caspase-3 level and activity. Whereas MERS-CoV infection potently induced caspase-3 expression and activity, both were restored to levels similar to that of mock infection control when Smad7 was knocked down (Fig. 3a). Separately, we administrated a small-molecule inhibitor, tyrphostin AG1296, which specifically blocks tyrosine kinase activity of the FGF receptor, to study the effect of FGF2 on MERS-CoV-induced apoptosis. Blockage of the downstream signalling of FGF2 dampened MERS-CoV-induced apoptosis by around 40% (Fig. 3b, top). However, control inhibitor tyrphostin AG490, which inhibits the epidermal growth factor receptor, failed to counteract MERS-CoV-induced apoptosis. The requirement of FGF2 for MERS-CoV-induced apoptosis was further validated by sequestration of secreted FGF2 using anti-FGF2 neutralizing antibodies. Addition of anti-FGF2 diminished caspase-3 expression to about 50% (Fig. 3b, bottom). Overall, our data consistently support the hypothesis that MERS-CoV induces a strong apoptotic response in both lung and kidney cells through ectopic expression of Smad7 and FGF2.
a, MERS-CoV-induced Smad7 expression in HK2 cells was dampened by transfection of siRNAs (Smad7-1 siRNA, lane 1; Smad7-2 siRNA, lane 2). Control siRNA-transfected (lane 3) and mock-infected (lane 4) cells were analysed. Left panels: protein expression of Smad7 and cleaved caspase-3. γ-Tubulin was detected as a loading control. Right panels: mRNA levels of Smad7 normalized to GAPDH and the relative apoptotic activities of the MERS-CoV-infected cells. b, FGF2 inhibition by a small-molecule compound or antibodies. Caspase-3 activities in MERS-CoV-infected HK2 cells were measured when FGF2 signalling was blocked with a small-molecule inhibitor AG1296 (top, lane 1). AG490 (top, lane 2), which blocks epidermal growth factor but not FGF2 signalling, and untreated MERS-CoV-infected cells (top, lane 3) served as negative and mock controls, respectively. Caspase-3 expression was determined in MERS-CoV-infected HK2 cells treated with anti-FGF2 neutralizing antibodies (bottom, lane 1) or irrelevant IgG (bottom, lane 2). Mock-infected cells were assessed (bottom, lane 3). γ-Tubulin was detected as the loading control. c, Cell protection by anti-Smad7 oligonucleotide against MERS-CoV-induced cytotoxicity in HK2 (dark diamonds) and Calu-3 (grey squares) cells when compared with that of untreated MERS-CoV infected cell lines. d, The antiviral activity of anti-Smad7 oligonucleotide was measured by comparing the levels of viral load from the MERS-CoV-infected cell extracts. All statistical significance was evaluated by Student's t-tests (P) and Pearson's correlation analyses (r). Images shown in a and b are representative of three independent experiments. Error bars in a–d represent the mean ± s.d. of three independent experiments. Samples from a–d represent biological replicates.
Given that the virus may promote apoptosis in infected cells in order to spread virus progeny to neighbouring cells, we hypothesized that apoptosis may be important for completion of the highly lytic MERS-CoV replication cycle. Although drugs specifically targeting Smad7 are currently unavailable, suppression of Smad7 expression using antisense oligonucleotides has been successfully applied in clinical trials to treat inflammatory bowel disease, with no major side effects19,20. To test whether an antisense oligonucleotide targeting Smad7 (anti-Smad7 oligo) could suppress MERS-CoV-induced apoptosis and inhibit virus replication, we treated both HK2 and Calu-3 cells with anti-Smad7 oligo before challenge with MERS-CoV. Intriguingly, cells treated with anti-Smad7 oligo showed a dose-dependent reduction in apoptosis. Cell protection assay by methylthiazolyldiphenyl-tetrazolium (MTT) showed that anti-Smad7 oligo protected 50% of cells from MERS-CoV-induced apoptosis at concentrations of 0.432 μg ml–1 and 0.965 μg ml–1 in HK2 and Calu-3 cells, respectively (Fig. 3c). Real-time quantitative polymerase chain reaction (RT-qPCR) revealed that virus production was inversely proportional to the concentration of the administrated anti-Smad7 oligo (Fig. 3d). These data were consistent with the results obtained from plaque assays that measured the amount of infectious virus. In anti-Smad7 oligo-treated HK2 cells, we observed a 1.4 log reduction in virus production (Supplementary Fig. 4). The strong protective effect of anti-Smad7 oligo against MERS-CoV-induced apoptosis and virus production makes it a potential treatment option for MERS. Furthermore, we determined the viral loads of culture supernatants, positive-strand and negative-strand viral RNAs in cell lysates and viral proteins at different time points in MERS-CoV infected HK2 cells with Smad7 or control siRNAs (Supplementary Fig. 5). The viral load of culture supernatant was significantly reduced with Smad7 siRNA at 12 hours post infection (h.p.i.), which suggested that Smad7 siRNA interfered with the step of virus release in the replication cycle. The important role of apoptosis on the viral replication cycle was further demonstrated by the treatment of infected HK2 cells by etoposide, an apoptosis-inducing drug, which increases virus production, and by MDIVI, an apoptosis inhibitor that suppresses virus production (Supplementary Fig. 6).
The host response to MERS-CoV infection was further investigated in a nonhuman primate model. We have previously established a rhesus macaque model21. More recently, another nonhuman primate model using common marmosets was established, which recapitulated the severe and sometimes lethal respiratory symptoms with disseminated extra-pulmonary infection seen in MERS patients22. We thus challenged a group of six common marmosets with MERS-CoV using our previous infection protocol22. We observed that all MERS-CoV-inoculated common marmosets developed ARDS, with one death due to severe illness (Supplementary Fig. 7). Successful infection was confirmed by the detection of viral RNA and antigen in all lung samples collected from MERS-CoV-inoculated common marmosets (Fig. 4a and Supplementary Fig. 8a,b). Consistent with the clinical findings in humans, a high viral load could be detected in the lungs of the infected animals, whereas no virus RNA could be detected in the control uninfected animal (Supplementary Fig. 8a). Histopathological examination of the lungs also revealed extensive broncho-interstitial pneumonia (Supplementary Fig. 8b,c). We further investigated the relationship of the abundance of MERS-CoV and the expression of Smad7 and FGF2. Remarkably, the results showed a good correlation of the amounts of MERS-CoV RNA with the expression levels of Smad7 and FGF2 (Fig. 4a and Supplementary Fig. 9). These findings were consistent with our ex vivo human organ culture and cell line models data, in which MERS-CoV mediates apoptosis through the induction of Smad7 and FGF2. Notably, viral RNA and antigen were also detected in four of six common marmosets' kidneys (Fig. 4b and Supplementary Fig. 8a). Intriguingly, the infected kidney samples displayed characteristic histological features of acute kidney injury with mitochrondrial shortening and fragmentation (AKI) (Fig. 4c, Supplementary Figs 10 and 11). While the development of AKI could be attributed partially to the septic shock of severe infection, the detection of viral RNA and antigen in the kidney samples (Fig. 4b, Supplementary Figs 8a and 11) raised the distinct possibility that MERS-CoV may cause direct damage to the infected kidneys. We also observed co-localization of MERS-CoV NP with caspase-3, Smad7 and FGF2 in renal tubular and lung epithelial cells, which was consistent with our cellular model in which MERS-CoV induced a direct cytopathic effect by apoptosis mediated through the upregulation of Smad7 and FGF2 (Fig. 4b, Supplementary Figs 2 and 8b). Taken together, our in vitro, ex vivo and in vivo results support the notion that Smad7 and FGF2 may play important roles in the pathogenesis of MERS-CoV-induced lung and kidney damage.
a, Quantitative measurement of MERS-CoV RNA in the lungs of MERS-CoV-inoculated common marmosets. Samples were divided into three categories (1.9 × 105 ± 9.9 × 104; 1.6 × 106 ± 5.2 × 105; 2.6 × 106 ± 1.7 × 106) based on the viral load (top). The relative expression levels of Smad7 (middle) and FGF2 (bottom) are indicated. All values were normalized to GAPDH. Statistical significance was evaluated by Student's t-tests and P values are indicated. Error bars represent the mean ± s.d. of three selected tissue samples. b, Co-immunohistochemical staining of MERS-CoV NP, caspase-3, Smad7 and FGF2 in kidneys of MERS-CoV-inoculated common marmosets. All co-immunohistochemical staining was performed on the same slide, except Smad7 and MERS-CoV NP, which were stained on separate slides, and the overlay view was generated using two neighbouring slides. The specificity of anti-MERS-CoV antibodies was confirmed by overnight pre-incubation with a fivefold more concentrated MERS-CoV NP peptide before their application to the kidney sections. The central image in the bottom panel represents the background signal detected after inoculation of rabbit secondary antibodies. Nuclei counterstained by DAPI are shown in blue. c, Haematoxylin and eosin staining of kidney sections of MERS-CoV-inoculated common marmosets. Interstitial infiltration was observed in the infected kidneys (arrow heads). Characteristic histological features of acute kidney injury, including flattened epithelial cells (arrows) and peritubular capillary congestion (white arrows), were detected in the kidney sections of MERS-CoV-inoculated common marmosets. Images shown in b and c are representatives of the four common marmosets' kidneys in which the MERS-CoV RNAs were detected (Supplementary Fig. 8a). D, dilated renal tubules; PC, protein cast.
Annual outbreaks of MERS in Middle East or other countries have been associated with a high mortality rate of over 30%, which is more than three times that of SARS2. Although both MERS and SARS patients died with multi-organ failure, MERS-CoV appears to have broader tissue tropism. We have previously demonstrated that MERS-CoV infection can induce apoptosis in T cells by activating both the extrinsic and intrinsic apoptosis pathways through an unknown mechanism23. Here, we show that the direct cytopathic effect of MERS-CoV is mediated through the induction of the expression of pro-apoptotic cellular proteins, Smad7 and FGF2, in infected lung and kidney cells. Recent work has demonstrated the pro-apoptotic role of Smad7 in hepatocellular carcinoma (HCC) through the attenuation of NFκB and TGFβ signalling16. These findings were in line with the idea that deregulated expression of Smad7 could be associated with renal pathogenesis12,13 through the induction of caspase 3-dependent14 and -independent apoptosis15 in a cell-type specific manner. Despite the fact that Smad7 plays important regulatory roles in TGFβ signalling16, distinct mechanisms of action have been proposed for Smad7- and TGFβ-induced apoptosis. It has been suggested that Smad7 mediates apoptosis through the inhibition of cell survival factor NF-κB, whereas TGFβ induces apoptosis by activation of the mitogen-activated protein (MAP) kinase p38. Overall, our results, together with previous reports, support the notion that the expression levels of both Smad7 and TGFβ should be tightly regulated. Different pathological conditions, including viral infection in different cell types, could abruptly alter the expression level of Smad7 or TGFβ, which may result in apoptosis. Meanwhile, FGF2 has been reported to induce apoptosis by altering the expression levels of apoptosis regulator and activating the mitogen-activated protein kinase (MAPK) pathway24. Further studies are warranted for the dissection of Smad7- and FGF2-induced apoptotic pathways on MERS-CoV infection. The unique cellular response of apoptosis induced by MERS-CoV, which results in virus release, may indicate an atypical mechanism of action for coronavirus replication. This highlights a possibility that the apoptotic process not only facilitates virus release and dissemination, but may also contribute to tissue damage, which may lead to ARDS and renal failure. These insights into the pathogenesis of MERS-associated organ damage may help to expand the limited anti-MERS treatment options identified by drug repurposing programmes or de novo development so far22,25–27. Potential intervention strategies counteracting Smad7 and FGF2 expression and/or signalling with various agents such as siRNAs, small-molecule inhibitors of FGF receptor tyrosine kinase and anti-Smad7 oligonucleotides may improve the outcome of MERS.
Methods
Viruses
The EMC/2012 strain of MERS-CoV (passage 8, designated MERS-CoV) was provided by R. Fouchier (Erasmus Medical Center)1 and SARS-CoV strain HKU39849 was propagated in VeroE6 cells (ATCC) in Dulbecco modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 100 units ml–1 penicillin plus 100 µg ml–1 streptomycin (1% PS). All experiments were performed as previously described according to biosafety level 3 practices26,28,29.
Virus titration by TCID50 assay
The 50% tissue culture infectious dose (TCID50) per ml was determined for MERS-CoV and SARS-CoV in VeroE6 cells as described previously6. Briefly, cells were plated in 96-well plates at a density of 5 × 104 cells per well in 150 µl DMEM. The virus was serially diluted by half-log from 103 to 1014 in DMEM. One hundred microlitres of each dilution was added per well and the plates were observed daily for cytopathic effect for five consecutive days.
Cell culture
Calu-3 (ATCC) is an epithelial cell line derived from human lung adenocarcinoma and were cultured in ATCC-formulated Eagle's Minimum Essential Medium (EMEM) supplemented with 10% FCS and 1% PS. Normal human mesangial cells (NHMCs; Lonza) were cultured in mesangial cell basal medium (MsBM; Lonza) supplemented with 10% FCS, 1% PS, 5 µg ml–1 transferrin and 5 µg ml–1 insulin. The podocyte cell line30 was cultured in RPMI (Life Technologies) supplemented with 10% FCS and 1% PS at 33 °C with 5% CO2. The podocytes were induced to differentiate at 37 °C for 3 days before infection. All experiments using NHMCs and podocytes were performed within their tenth passage. All cell lines used in the study were authenticated by Southern and FISH analyses and were confirmed to be free of mycoplasma contamination as determined by Plasmo Test (InvivoGen).
Reagents and overexpression constructs
Etoposide and MDIVI were purchased from Cell Signaling Technology and Sigma, respectively. The overexpression construct of BIRC3 was purchased from Origene. siGENOME human siRNA-SMARTpools targeting CLU, SOD1 and CAT and Smad7-1 siRNA (5′-GUUCAGGACCAAACGAUCUGC-3′; 5′-GCAGAUCGUUUGGUCCUGAACAU-3′) and Smad7-2 siRNA (5′-CUCACGCACUCGGUGCUCAAG-3′; 5′-CUUGAGCACCGAGUGCGUGAGCG-3′) were purchased from Thermo Scientific Dharmacon. Antisense oligonucleotide targeting Smad7 (anti-Smad7 oligo; 5′-GTCGCCCCTTCTCCCCGCAG-3′; also named GED0301)19,20 was synthesized by Integrated DNA Technologies with the following modifications: (1) the cytosine preceding the guanosine was replaced with a 5-methyl-deoxycytosine and (2) the internucleotide linkages of the backbone were modified to O,O-linked phosphorothioates.
Ex vivo human kidney and lung organ culture for MERS-CoV infection
All experiments involving human tissues were approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster. Fresh surgically resected kidney or lung specimens were obtained from patients undergoing nephrectomy or pneumonectomy, respectively, at Queen Mary Hospital, Hong Kong, as part of standard clinical management. Ex vivo organ cultures were carried out as previously described28. The kidney cortical and lung tissues (in small strips, 1 mm width) were placed directly into a 24-well plate with 1 ml of Kaighn's Modification of Ham's F-12 Medium (F-12 K) with 1% PS at 37 °C. Tissues were infected with MERS-CoV or SARS-CoV at a virus concentration of 1 × 106 TCID50 per ml for 4 h at 37 °C. Unbound viruses were washed away with phosphate buffered saline (PBS). Tissues were collected at 0, 18, 48 and 72 h.p.i. and then fixed in 10% paraformaldehyde for immunohistochemistry and electron microscopy.
Common marmoset infection model
All experiments involving common marmosets were performed as previously described and were approved by the Institutional Animal Care and Use Committee21. Briefly, six male common marmosets (Callithrix jacchus; 3 years old) were inoculated with 5 × 106 TCID50 of MERS-CoV intratracheally in 500 µl DMEM. Sample sizes were based on the availability of common marmosets. Necropsies of the six common marmosets were scheduled for 3 days post infection (d.p.i.). The common marmosets were observed twice daily for clinical signs of disease and scored using a clinical scoring system prepared for common marmosets as described22 (see Supplementary Section ‘Berlin definition of ARDS’). On 1 and 3 d.p.i., clinical examinations and X-rays were performed on anaesthetized animals. The organs were collected on necropsy for experimental assays as described in ref. 21. No randomization or blinding was done in this study.
Immunohistochemical staining and electron microscopy
Infected primate kidneys were fixed in 10% paraformaldehyde and cryosectioned. The mounted slides were blocked with TBS4 for 1 h followed by 0.1% Sudan Black B (Sigma) for another 25 min. Excess Sudan Black B was washed away three times with PBS and Tween-20 (PBST). MERS-CoV NP was stained with mouse anti-MERS-CoV NP (1:200) overnight at 4 °C as previously described28. Rabbit antibodies against cell type-specific markers synaptopodin (Abcam), CD31 (Abcam), cytokeratin 18 (Abcam), Smad7 (Santa Cruz), FGF2 (Abcam) and caspase-3 (Cell Signaling Technology) were also used. Unbound antibodies were washed away three times with PBST. Alexa Fluor 488 goat anti-mouse IgG (H+L) antibodies (1:500; Life Technologies) and Alexa Fluor 594 goat anti-rabbit IgG (1:500; Life Technologies) were then applied to the slides for signal detection, followed by washing three times with PBST. Stained slides were mounted with coverslips in the presence of anti-fade mountant (Dako) and 1 µg ml–1 4′,6-diamidino-2-phenylindole (DAPI) (Sigma). Human kidney tissues were embedded in Tissue-Tek OCT compound (Miles), snap frozen and stored at −80 °C before sectioning. Sections (5–10 µm) were cut on a cryostat and transferred to glass microscope slides pre-coated with poly-lysine (Sigma). Sections were then fixed with cold acetone for 20 min at room temperature. Fixed sections were washed three times with PBS before blocking and staining with anti-MERS-CoV NP as described above. Digital images were acquired using NIKON Eclipse Ni-U with SPOT RT3 camera. To prepare samples for electron microscopic examination, the paraformaldehyde-fixed tissues were post-fixed for 30 min with 0.5% osmium tetroxide/0.8% potassium ferricyanide in 0.1 M sodium cacodylate, for 1 h with 1% tannic acid and overnight with 1% uranyl acetate at 4 °C. Specimens were dehydrated with a graded ethanol series from water and then successively at 10, 20, 50, 95 and 100% ethanol followed by two final exchanges in 100% propylene oxide before infiltration and final embedding in Embed-812 resin. Thin sections were cut with a Leica EM UC6 μLtramicrotome (Leica) and stained with 1% uranyl acetate and Reynold's lead citrate before viewing at 120 kV on a FEI Tecnai G2 20 S-TWIN scanning transmission electron microscope. Digital images were acquired with a Gatan 794 1K × 1K Camera System with DigitalMicrograph Acquisition Software.
Reverse transcription and quantitative real-time PCR (RT-qPCR)
Total RNAs and viral RNAs were isolated using Trizol (Life Technologies) and the Viral RNA Mini kit (QIAGEN), respectively, as previously described6. After RNA quantification, 1 µg of RNAs was reverse transcribed into cDNA using random hexamers. Total MERS-CoV transcripts were detected with a Novel Coronavirus 2012 Real-Time RT-PCR assay (CDC; catalog no. KT0136). The viral RNAs isolated from cell lysates and conditioned media were normalized to the mRNA expression level of GAPDH and a spike-in control, enterovirus 71 (EV71) strain SZ/HK08-5, using primers 5′-GCTCACTGGCATGGCCTTCCGTGT-3′ and 5′-TGGAGGAGTGGGTGTCGCTGTTGA-3′ (GAPDH) as well as 5′-CCCCTGAATGCGGCTAATCC-3′ and 5′- ACACGGACACCCAAAGTAGT-3′ (EV71). For each reaction, equal amounts of cDNA were mixed with FS Universal SYBR Green Master Rox (Roche) plus 5 pmol each of forward and reverse primers. Amplification was performed for 15 s at 95 °C and 1 min at 60 °C for 55 cycles in a 7900 real-time PCR detection system (ABI).
High-throughput sequencing
Multiple dishes of cells were seeded under the same culture conditions. Infections of all dishes of cells were performed simultaneously using a master mix of MERS-CoV or SARS-CoV to minimize variation. At the indicated time points, total RNAs of the infected cells were collected. The altered gene expression following MERS-CoV or SARS-CoV infection at different time points were then compared with those at time point zero. The samples from time point zero were subjected to the corresponding virus inoculation, then immediately washed and immediately harvested to ensure that the detected gene alteration was not due to the virus inoculation process. The sequencing libraries were prepared at the Centre for Genomic Sciences at the University of Hong Kong using the TruSeq Stranded mRNA Sample Prep Kit (Illumina) and were subsequently sequenced on an Illumina HiSeq 2000. Briefly, mRNA was isolated using polyT oligo-attached magnetic beads. The purified mRNA was fragmented and reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen) to first-strand cDNA in the presence of random hexamers. Double-stranded cDNAs were synthesized by second strand synthesis and then subjected to end repair, 3′ adenylation and indexed adaptor ligation. The adaptor-ligated sequencing libraries were further enriched by ten cycles of PCR, quantified and sequenced on a HiSeq 2000 with 101 bp paired-end reads. Image analyses and base calling were performed with SCS2.8/RTA1.8 (Illumina). FASTQ file generation and the removal of failed reads were performed using CASAVA ver.1.8.2 (Illumina). Pathway analyses were performed using the Ingenuity Pathway Analysis software. Candidate genes from a defined disease pathway (renal necrosis/cell death) that possessed the following criteria were selected for further analyses: (1) the absolute gene expression level should be markedly induced; (2) the trend of gene expression should be consistent throughout the course of infection; (3) the tissue-specificity of the identified genes should not be restricted only to lung, but should be expressed in kidney; (4) a differential response of the gene expression pattern should be observed when infected with MERS-CoV and SARS-CoV, separately.
Apoptosis assays
A caspase-3/CPP32 fluorometric assay kit (BioVision) was used to measure caspase-3 activity. Briefly, cells were plated in six-well dishes, cultured, collected and lysed in lysis buffer at 0, 8, 24, 48 and 72 h.p.i. After the protein concentration was normalized by the Bradford assay, the lysates were incubated with the same amounts of reaction buffer and 50 mmol l–1 DEVD-AFC substrate for 2 h at 37 °C. Fluorescence was monitored with an excitation wavelength of 400 nm and an emission wavelength of 505 nm. Apoptosis in cells that were infected by MERS-CoV or SARS-CoV or transfected with overexpression constructs Smad7 (Origene) or FGF2 (Origene) were measured by ApopTag Red In Situ Apoptosis Detection Kit (Chemicon). Briefly, treated cells on the coverslips were fixed with 1% paraformaldehyde. Cells undergoing apoptosis with DNA breakage will be labelled with TdT enzyme in the presence of digoxigennin-modified nucleotides. The coverslips were then mounted on the slides in the presence of VECTASHIELD Antifade mounting medium with DAPI (Vector lab). Signal detection was facilitated by the addition of anti-digoxigenin conjugate followed by examination under a fluorescence microscopy (NIKON Eclipse Ni-U) with SPOT RT3 camera.
Accession numbers
Data have been deposited in the Sequence Read Archive under accession code SRP056612.
References
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012).
Chan, J. F. et al. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 28, 465–522 (2015).
Arabi, Y. M. et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann. Intern. Med. 160, 389–397 (2014).
Chan, J. F. et al. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 65, 477–489 (2012).
Eckerle, I., Muller, M. A., Kallies, S., Gotthardt, D. N. & Drosten, C. In-vitro renal epithelial cell infection reveals a viral kidney tropism as a potential mechanism for acute renal failure during Middle East Respiratory Syndrome (MERS) Coronavirus infection. Virol. J. 10, 359 (2013).
Chan, J. F. et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 207, 1743–1752 (2013).
Chu, K. H. et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 67, 698–705 (2005).
Hung, I. F. et al. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 10, 1550–1557 (2004).
Poissy, J. et al. Kinetics and pattern of viral excretion in biological specimens of two MERS-CoV cases. J. Clin. Virol. 61, 275–278 (2014).
Drosten, C. et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 13, 745–751 (2013).
Ito, H., Bando, H., Shimada, T. & Katsuma, S. The BIR and BIR-like domains of Bombyx mori nucleopolyhedrovirus IAP2 protein are required for efficient viral propagation. Biochem. Biophys. Res. Commun. 454, 581–587 (2014).
Schiffer, M. et al. Apoptosis in podocytes induced by TGF-beta and Smad7. J. Clin. Invest. 108, 807–816 (2001).
Lan, H. Y. et al. Inhibition of renal fibrosis by gene transfer of inducible Smad7 using ultrasound-microbubble system in rat UUO model. J. Am. Soc. Nephrol. 14, 1535–1548 (2003).
Okado, T. et al. Smad7 mediates transforming growth factor-beta-induced apoptosis in mesangial cells. Kidney Int. 62, 1178–1186 (2002).
Bitzer, M. et al. A mechanism of suppression of TGF-β/SMAD signaling by NF-κB/RelA. Genes Dev. 14, 187–197 (2000).
Wang, J. et al. Inhibitory role of Smad7 in hepatocarcinogenesis in mice and in vitro. J. Pathol. 230, 441–452 (2013).
Floege, J. et al. Basic fibroblast growth factor augments podocyte injury and induces glomerulosclerosis in rats with experimental membranous nephropathy. J. Clin. Invest. 96, 2809–2819 (1995).
Li, Z., Jerebtsova, M., Liu, X. H., Tang, P. & Ray, P. E. Novel cystogenic role of basic fibroblast growth factor in developing rodent kidneys. Am. J. Physiol. Renal Physiol. 291, F289–F296 (2006).
Zorzi, F. et al. A phase 1 open-label trial shows that smad7 antisense oligonucleotide (GED0301) does not increase the risk of small bowel strictures in Crohn's disease. Aliment. Pharmacol. Ther. 36, 850–857 (2012).
Monteleone, G. et al. Phase I clinical trial of Smad7 knockdown using antisense oligonucleotide in patients with active Crohn's disease. Mol. Ther. 20, 870–876 (2012).
Yao, Y. et al. An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J. Infect. Dis. 209, 236–242 (2014).
Chan, J. F. et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 212, 1904–1913 (2015).
Chu, H. et al. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. http://dx.doi.org/10.1093/infdis/jiv380 (2015).
Matsumoto, T., Turesson, I., Book, M., Gerwins, P. & Claesson-Welsh, L. p38 MAP kinase negatively regulates endothelial cell survival, proliferation, and differentiation in FGF-2-stimulated angiogenesis. J. Cell. Biol. 156, 149–160 (2002).
Chan, J. F. et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 67, 606–616 (2013).
Lu, L. et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nature Commun. 5, 3067 (2014).
Zumla, A . et al. Coronaviruses—drug discovery and therapeutic options. Nature Rev. Drug Discov. (in the press).
Zhou, J. et al. Active MERS-CoV replication and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 209, 1331–1342 (2014).
Chu, H. et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology 454–455, 197–205 (2014).
Ni, L., Saleem, M. & Mathieson, P. W. Podocyte culture: tricks of the trade. Nephrology (Carlton) 17, 525–531 (2012).
Acknowledgements
This work was supported in part by donations from the Shaw Foundation, R. Yu and C. Yu, the Providence Foundation (in memory of the late Lui Hac Minh), Cheer Master Investments, and Respiratory Viral Research Foundation Limited; and funding from Seed Funding for the Theme-based Research Scheme and Strategic Research Theme Fund from the University of Hong Kong, the Hong Kong Health and Medical Research Fund (13121102, 14130822, 14131392 and HKM-15-M01), the Hong Kong Research Grants Council (N_HKU728/14, HKU1/CRF/11G and 17124415) and the National Science and Technology Major Projects of Infectious Disease (2012ZX10004501-004). The authors thank A. Ng, C. Lau, L.-K. Tang and members of the Centre for Genomic Sciences, The University of Hong Kong, for their technical support.
Author information
Authors and Affiliations
Contributions
M.-L.Y. and K.-Y.Y. conceived and designed the study. M.-L.Y. and L.J. performed most experiments with the help of K.-H.C., J.L.L.T., H.C. and J.Z. M.-L.Y. and A.K.L.T. performed bioinformatics analysis. Y.Y., J.F.W.C., V.K.M.P., W.D., B.-J.Z. and C.Q. constructed the animal model, provided samples and analysed the data. K.-F.C., Q.Z., T.-M.C. and S.Y. performed histopathological analysis. J.F.W.C., K.K.W.T. and M.-K.Y. provided samples and assisted with the establishment of ex vivo organ culture. S.K.P.L. and P.C.Y.W. provided advice, assisted with sequencing and analysed the data. M.-L.Y., C.Q. and K.-Y.Y. wrote the manuscript. Y.Y., J.F.W.C., H.C., J.L.L.T., J.Y.N.L., D.-Y.J. and P.W.M. provided advice and contributed to data analysis and manuscript preparation. C.Q. and K.-Y.Y. secured funding and conducted trouble-shooting. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–12, References, Definitions and Table 1 (PDF 4620 kb)
Rights and permissions
About this article
Cite this article
Yeung, ML., Yao, Y., Jia, L. et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol 1, 16004 (2016). https://doi.org/10.1038/nmicrobiol.2016.4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.4
This article is cited by
-
Nanostructures for prevention, diagnosis, and treatment of viral respiratory infections: from influenza virus to SARS-CoV-2 variants
Journal of Nanobiotechnology (2023)
-
Inflammatory cell death, PANoptosis, screen identifies host factors in coronavirus innate immune response as therapeutic targets
Communications Biology (2023)
-
Proviral role of caspase-6 in coronavirus infections
Cell Research (2022)
-
A comprehensive mini-review on amyloidogenesis of different SARS-CoV-2 proteins and its effect on amyloid formation in various host proteins
3 Biotech (2022)
-
Coronaviruses exploit a host cysteine-aspartic protease for replication
Nature (2022)