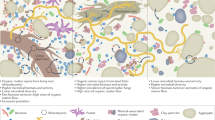
Abstract
Extracellular DNA from dead microorganisms can persist in soil for weeks to years1–3. Although it is implicitly assumed that the microbial DNA recovered from soil predominantly represents intact cells, it is unclear how extracellular DNA affects molecular analyses of microbial diversity. We examined a wide range of soils using viability PCR based on the photoreactive DNA-intercalating dye propidium monoazide4. We found that, on average, 40% of both prokaryotic and fungal DNA was extracellular or from cells that were no longer intact. Extracellular DNA inflated the observed prokaryotic and fungal richness by up to 55% and caused significant misestimation of taxon relative abundances, including the relative abundances of taxa integral to key ecosystem processes. Extracellular DNA was not found in measurable amounts in all soils; it was more likely to be present in soils with low exchangeable base cation concentrations, and the effect of its removal on microbial community structure was more profound in high-pH soils. Together, these findings imply that this ‘relic DNA’ remaining in soil after cell death can obscure treatment effects, spatiotemporal patterns and relationships between microbial taxa and environmental conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
14 July 2017
In the PDF version of this article previously published, the year of publication provided in the footer of each page and in the 'How to cite' section was erroneously given as 2017, it should have been 2016. This error has now been corrected. The HTML version of the article was not affected.
References
Nielsen, K. M., Johnsen, P. J., Bensasson, D. & Daffonchio, D. Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6, 37–53 (2007).
Pietramellara, G. et al. Extracellular DNA in soil and sediment: fate and ecological relevance. Biol. Fertil. Soils 45, 219–235 (2009).
Levy-Booth, D. J. et al. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 39, 2977–2991 (2007).
Nocker, A., Sossa-Fernandez, P., Burr, M. D. & Camper, A. K. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 73, 5111–5117 (2007).
Roesch, L. F. W. et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1, 283–290 (2007).
Gans, J., Wolinsky, M. & Dunbar, J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, 1387–1390 (2005).
Fierer, N. et al. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73, 7059–7066 (2007).
Brown, C. T. et al. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523, 208–211 (2015).
Jones, M. D. M. et al. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474, 200–203 (2011).
Mackelprang, R. et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480, 368–371 (2011).
Ogram, A., Sayler, G. S., Gustin, D. & Lewis, R. J. DNA adsorption to soils and sediments. Environ. Sci. Technol. 22, 982–984 (1988).
Saeki, K. & Kunito, T. in Current Research Technology and Education Topics in Applied Microbiology and Microbial Biotechnology Vol. 1 (ed. Méndez-Vilas, A. ) 188–195 (Formatex Research Center, 2010).
Nielsen, K. M., Calamai, L. & Pietramellara, G . in Nucleic Acids and Proteins in Soil (eds Nannipieri, P. & Smalla, K. ) Ch. 7, 141–157 (Springer, 2006).
Agnelli, A. et al. Distribution of microbial communities in a forest soil profile investigated by microbial biomass, soil respiration and DGGE of total and extracellular DNA. Soil Biol. Biochem. 36, 859–868 (2004).
Dlott, G., Maul, J. E., Buyer, J. & Yarwood, S. Microbial rRNA:rDNA gene ratios may be unexpectedly low due to extracellular DNA preservation in soils. J. Microbiol. Methods 115, 112–120 (2015).
Nocker, A., Cheung, C.-Y. & Camper, A. K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320 (2006).
Cangelosi, G. A. & Meschke, J. S. Dead or alive: molecular assessment of microbial viability. Appl. Environ. Microbiol. 80, 5884–5891 (2014).
Fittipaldi, M., Nocker, A. & Codony, F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 91, 276–289 (2012).
Nocker, A., Richter-Heitmann, T., Montijn, R., Schuren, F. & Kort, R. Discrimination between live and dead cellsin bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int. Microbiol. 13, 59–65 (2010).
Wagner, A. O., Praeg, N., Reitschuler, C., Illmer, P. & Illmer, P. Effect of DNA extraction procedure, repeated extraction and ethidium monoazide (EMA)/propidium monoazide (PMA) treatment on overall DNA yield and impact on microbial fingerprints for bacteria, fungi and archaea in a reference soil. Appl. Soil Ecol. 93, 56–64 (2015).
Rocca, J. D. et al. Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J. 9, 1693–1699 (2015).
Wertz, S., Leigh, A. K. K. & Grayston, S. J. Effects of long-term fertilization of forest soils on potential nitrification and on the abundance and community structure of ammonia oxidizers and nitrite oxidizers. FEMS Microbiol. Ecol. 79, 142–154 (2012).
Bolan, N. S. A critical review on the role of mycorrhizal fungi in the uptake of phosphorus by plants. Plant Soil 134, 189–207 (1991).
Tedersoo, L. et al. Global diversity and geography of soil fungi. Science 346, 1256688 (2014).
Lauber, C. L., Hamady, M., Knight, R. & Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Griffiths, R. I. et al. The bacterial biogeography of British soils. Environ. Microbiol. 13, 1642–1654 (2011).
Zhang, X., Barberán, A., Zhu, X., Zhang, G. & Han, X. Water content differences have stronger effects than plant functional groups on soil bacteria in a steppe ecosystem. PLoS ONE 9, e115798 (2014).
Docherty, K. M. et al. Key edaphic properties largely explain temporal and geographic variation in soil microbial communities across four biomes. PLoS ONE 10, e0135352 (2015).
Bond-Lamberty, B. et al. Soil respiration and bacterial structure and function after 17 years of a reciprocal soil transplant experiment. PLoS ONE 11, e0150599 (2016).
Howe, A. C. et al. Tackling soil diversity with the assembly of large, complex metagenomes. Proc. Natl Acad. Sci. USA 111, 4904–4909 (2014).
Ramirez, K. S. et al. Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc. R. Soc. B 281, 20141988 (2014).
Dell'Anno, A. & Danovaro, R. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309, 2179–2179 (2005).
Coolen, M. J. L. et al. Evolution of the plankton paleome in the Black Sea from the Deglacial to Anthropocene. Proc. Natl Acad. Sci. USA 110, 8609–8614 (2013).
Inagaki, F., Okada, H., Tsapin, A. I. & Nealson, K. H. Microbial survival: the paleome: a sedimentary genetic record of past microbial communities. Astrobiology 5, 141–153 (2005).
Kemper, W. D. & Rosenau, R. C. in Methods of Soil Analysis: Part 1—Physical and Mineralogical Methods (ed. Klute, A. ) 425–442 (Soil Science Society of America, 1986).
Barbau-Piednoir, E. et al. Evaluation of viability-qPCR detection system on viable and dead Salmonella serovar Enteritidis. J. Microbiol. Methods 103, 131–137 (2014).
Pinheiro, E. T., Neves, V. D., Reis, C. C., Longo, P. L. & Mayer, M. P. Evaluation of the propidium monoazide–quantitative polymerase chain reaction method for the detection of viable Enterococcus faecalis. J. Endod. 42, 1089–1092 (2016).
Taylor, M. J., Bentham, R. H. & Ross, K. E. Limitations of using propidium monoazide with qPCR to discriminate between live and dead Legionella in biofilm samples. Microbiol. Insights 2014, 15–24 (2014).
Blazewicz, S. J., Barnard, R. L., Daly, R. A. & Firestone, M. K. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068 (2013).
Durfee, T. et al. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 190, 2597–2606 (2008).
Herrera, M. L., Vallor, A. C., Gelfond, J. A., Patterson, T. F. & Wickes, B. L. Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J. Clin. Microbiol. 47, 1325–1332 (2009).
Leff, J. W. et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl Acad. Sci. USA 112, 10967–10972 (2015).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
McDonald, D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2011).
Abarenkov, K. et al. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol. 186, 281–285 (2010).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Leff, J. W. mctoolsr: Microbial Community Data Analysis Tools. v.0.1.0.14 (2016); http://leffj.github.io/mctoolsr/
Storey, J. D. A direct approach to false discovery rates. J. R. Stat. Soc. B 64, 479–498 (2002).
Harrell, F. E. Jr & Dupont, C. Hmisc: Harrell Miscellaneous. v.4.0-0 (2016); https://cran.r-project.org/web/packages/Hmisc/index.html
Acknowledgements
The authors thank S. Grandy and J. Schnecker at the University of New Hampshire, C. Bueno de Mesquita and L. Vimercati at the University of Colorado Boulder, E. Skokan at Black Cat Farms and K. McLachlan at Kansas State University for soil collection or access to collection sites. The authors also thank J. Henley, R. Hacker-Cary and K. Vaccarello for assistance with DNA extractions. Sequencing was performed at the University of Colorado BioFrontiers Institute's Next-Gen Sequencing Core Facility. Funding to support this work was provided by the National Science Foundation (DEB 0953331, EAR 1331828, DUE 1259336 and EAR 1461281) and a Visiting Postdoctoral Fellowship award to P.C. from the Cooperative Institute for Research in Environmental Sciences.
Author information
Authors and Affiliations
Contributions
P.C. and N.F. conceived the project and wrote the manuscript. P.C., P.J.M. and E.E.M. performed experiments. P.C., P.J.M., N.F. and M.S.S. collected samples. P.C., J.W.L. and M.S.S. analysed data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-11. (PDF 950 kb)
Supplementary Dataset 1
Full dataset for Fig. 1; mean percent of each taxa per soil, per treatment; and amplicon copies per replicate per gram of soil. (XLSX 90 kb)
Supplementary Table 1
List of soil sample locations, ecosystem descriptions, edaphic characteristics, mean community dissimilarity after relic DNA removal, mean percent of relic DNA for each soil, mean richness, and mean 16S and ITS amplicon abundances for both untreated and PMA-treated soils. (XLSX 56 kb)
Rights and permissions
About this article
Cite this article
Carini, P., Marsden, P., Leff, J. et al. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol 2, 16242 (2017). https://doi.org/10.1038/nmicrobiol.2016.242
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.242
This article is cited by
-
Benchmarking bioinformatic virus identification tools using real-world metagenomic data across biomes
Genome Biology (2024)
-
Synthetic microbiology in sustainability applications
Nature Reviews Microbiology (2024)
-
Biotic homogenization, lower soil fungal diversity and fewer rare taxa in arable soils across Europe
Nature Communications (2024)
-
Effect of soil depth on the structure of bacterial composition in the active layer at five geologically distinct sites on James Ross and Vega Islands in Antarctica
Polar Biology (2024)
-
A hitchhiker’s guide: estimates of microbial biomass and microbial gene abundance in soil
Biology and Fertility of Soils (2024)