Abstract
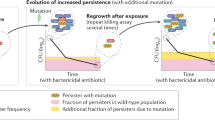
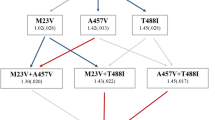
The evolution of antibiotic resistance is a major threat to society and has been predicted to lead to 10 million casualties annually by 20501. Further aggravating the problem, multidrug tolerance in bacteria not only relies on the build-up of resistance mutations, but also on some cells epigenetically switching to a non–growing antibiotic-tolerant ‘persister’ state2–6. Yet, despite its importance, we know little of how persistence evolves in the face of antibiotic treatment7. Our evolution experiments in Escherichia coli demonstrate that extremely high levels of multidrug tolerance (20–100%) are achieved by single point mutations in one of several genes and readily emerge under conditions approximating clinical, once-daily dosing schemes. In contrast, reversion to low persistence in the absence of antibiotic treatment is relatively slow and only partially effective. Moreover, and in support of previous mathematical models8–10, we show that bacterial persistence quickly adapts to drug treatment frequency and that the observed rates of switching to the persister state can be understood in the context of ‘bet-hedging’ theory. We conclude that persistence is a major component of the evolutionary response to antibiotics that urgently needs to be considered in both diagnostic testing and treatment design in the battle against multidrug tolerance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Review on Antimicrobial Resistance. Tackling a Crisis for the Health and Wealth of Nations (2015); http://amr-review.org/sites/default/files/Report-52.15.pdf
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Fauvart, M., De Groote, V. N. & Michiels, J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 60, 699–709 (2011).
Maisonneuve, E. & Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014).
Verstraeten, N. et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21 (2015).
Balaban, N. Q., Gerdes, K., Lewis, K. & McKinney, J. D. A problem of persistence: still more questions than answers? Nature Rev. Microbiol. 11, 587–591 (2013).
Kussell, E., Kishony, R., Balaban, N. Q. & Leibler, S. Bacterial persistence: a model of survival in changing environments. Genetics 169, 1807–1814 (2005).
Gardner, A., West, S. A. & Griffin, A. S. Is bacterial persistence a social trait? PLoS ONE 2, e752 (2007).
Patra, P. & Klumpp, S. Population dynamics of bacterial persistence. PLoS ONE 8, e62814 (2013).
Balázsi, G., Van Oudenaarden, A. & Collins, J. J. Cellular decision making and biological noise: from microbes to mammals. Cell 144, 910–925 (2011).
Veening, J.-W., Smits, W. K. & Kuipers, O. P. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210 (2008).
Kint, C. I., Verstraeten, N., Fauvart, M. & Michiels, J. New-found fundamentals of bacterial persistence. Trends Microbiol. 20, 577–585 (2012).
Stepanyan, K. et al. Fitness trade-offs explain low levels of persister cells in the opportunistic pathogen Pseudomonas aeruginosa. Mol. Ecol. 24, 1572–1583 (2015).
Mulcahy, L. R., Burns, J. L., Lory, S. & Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192, 6191–6199 (2010).
Fridman, O., Goldberg, A., Ronin, I., Shoresh, N. & Balaban, N. Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014).
Lee, H. H., Molla, M. N., Cantor, C. R. & Collins, J. J. Bacterial charity work leads to population-wide resistance. Nature 467, 82–85 (2010).
Allison, K. R., Brynildsen, M. P. & Collins, J. J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220 (2011).
Wakamoto, Y. et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339, 91–95 (2013).
Orman, M. A. & Brynildsen, M. P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob. Agents Chemother. 57, 3230–3239 (2013).
Acar, M., Mettetal, J. T. & van Oudenaarden, A. Stochastic switching as a survival strategy in fluctuating environments. Nature Genet. 40, 471–475 (2008).
Philippi, T. & Seger, J. Hedging one's evolutionary bets, revisited. Trends Ecol. Evol. 4, 41–44 (1989).
Baskin, C. C. & Baskin, J. M. Seeds: ecology, biogeography, and evolution of dormancy and germination (Elsevier, 2014).
Beaumont, H. J. E., Gallie, J., Kost, C., Ferguson, G. C. & Rainey, P. B. Experimental evolution of bet hedging. Nature 462, 90–93 (2009).
Kussell, E. & Leibler, S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078 (2005).
McKenzie, C. Antibiotic dosing in critical illness. J. Antimicrob. Chemother. 66, ii25–ii31 (2011).
ABC Project Team Ascertaining Barriers for Compliance: Policies for Safe, Effective and Cost-effective use of Medicines in Europe (2012); http://abcproject.eu/img/ABCFinal.pdf
Oz, T. et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol. Biol. Evol. 31, 2387–2401 (2014).
Andersson, D. I. & Hughes, D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nature Rev. Microbiol. 8, 260–271 (2010).
Cohen, N. R., Lobritz, M. A. & Collins, J. J. Microbial persistence and the road to drug resistance. Cell Host Microbe 13, 632–642 (2013).
Yu, J., Xiao, J., Ren, X., Lao, K. & Xie, X. S. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006).
Cherepanov, P. P. & Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995).
Fux, C. A., Costerton, J. W., Stewart, P. S. & Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40 (2005).
Eng, R. H. K., Padberg, F. T., Smith, S. M., Tan, E. N. & Cherubin, C. E. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35, 1824–1828 (1991).
Wu, M.-L., Tan, J. & Dick, T. Eagle effect in non-replicating persister Mycobacteria. Antimicrob. Agents Chemother. 59, 7786–7789 (2015).
Gocke, E. Mechanism of quinolone mutagenicity in bacteria. Mutat. Res. 248, 135–143 (1991).
Rodríguez-Rojas, A., Rodríguez-Beltrán, J., Couce, A. & Blázquez, J. Antibiotics and antibiotic resistance: A bitter fight against evolution. Int. J. Med. Microbiol. 303, 293–297 (2013).
Lázár, V. et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nature Commun. 5, 4352 (2014).
Ruiz, J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51, 1109–1117 (2003).
Kim, S., Lieberman, T. D. & Kishony, R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl Acad. Sci. 111, 14494–14499 (2014).
Wiser, M. J., Ribeck, N. & Lenski, R. E. Long-term dynamics of adaptation in asexual populations. Science 1364, 1364–1367 (2013).
Wiegand, I., Hilpert, K. & Hancock, R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protoc. 3, 163–175 (2008).
Liebens, V. et al. Identification and characterization of an anti-pseudomonal dichlorocarbazol derivative displaying anti-biofilm activity. Bioorg. Med. Chem. Lett. 24, 5404–5408 (2014).
Thierauf, A., Perez, G. & Maloy, A. S. Generalized transduction. Methods Mol. Biol. 501, 267–286 (2009).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Otto, S. P. & Day, T. A Biologist's Guide to Mathematical Modeling in Ecology and Evolution (Princeton University Press, 2007).
Helaine, S. et al. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl Acad. Sci. 107, 3746–3751 (2010).
Francois, K. et al. Modelling the individual cell lag phase. Isolating single cells: protocol development . Lett. Appl. Microbiol. 37, 26–30 (2003).
Acknowledgements
B.V.D.B. is a Research Foundation - Flanders (FWO)-fellow and J.E.M. is a fellow of the Agency for Innovation by Science and Technology (IWT). The research was supported by the KU Leuven Research Council (PF/10/010; PF/10/07; IDO/09/010; IDO/13/008; CREA/13/019; DBOF/12/035; DBOF/14/049), Interuniversity Attraction Poles–Belgian Science Policy Office (IAP-BELSPO) (IAP P7/28), ERC (241426), Human Frontier Science Program (HFSP) (RGP0050/2013), FWO (G047112N, KAN2014 1.5.222.14), Flanders Institute for Biotechnology(VIB) and the European Molecular Biology organisation (EMBO). We thank S. Xie for providing the ancestor strains and E. Toprak and S. Diggle for their comments on this manuscript.
Author information
Authors and Affiliations
Contributions
B.V.D.B. designed and performed the experiments, analysed the data and wrote the manuscript. J.E.M. helped in performing the experiments and analysing the data and edited the manuscript. T.W. made the evolutionary model, helped in analysing the data and edited the manuscript. E.M.W. helped in analysing the evolutionary model. P.V.B. and D.K. helped in performing the experiments. L.D.M. and K.J.V. edited the manuscript. N.V., M.F. and J.M. designed the experiments and edited the manuscript. J.E.M. and T.W. contributed equally. M.F. and J.M. contributed equally as senior authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1-8, Tables 1 and 2, Methods and References. (PDF 1311 kb)
Rights and permissions
About this article
Cite this article
Van den Bergh, B., Michiels, J., Wenseleers, T. et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol 1, 16020 (2016). https://doi.org/10.1038/nmicrobiol.2016.20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.20
This article is cited by
-
Deep mutational scanning of essential bacterial proteins can guide antibiotic development
Nature Communications (2023)
-
Bacterial defences: mechanisms, evolution and antimicrobial resistance
Nature Reviews Microbiology (2023)
-
Intermittent antibiotic treatment of bacterial biofilms favors the rapid evolution of resistance
Communications Biology (2023)
-
Systematic analyses identify modes of action of ten clinically relevant biocides and antibiotic antagonism in Acinetobacter baumannii
Nature Microbiology (2023)
-
Proteomic analysis of the initial wake up of vibrio splendidus persister cells
World Journal of Microbiology and Biotechnology (2023)