Abstract
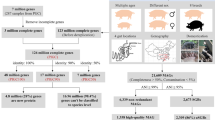
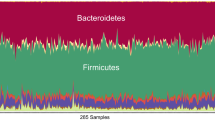
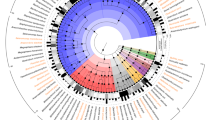
The pig is a major species for livestock production and is also extensively used as the preferred model species for analyses of a wide range of human physiological functions and diseases1. The importance of the gut microbiota in complementing the physiology and genome of the host is now well recognized2. Knowledge of the functional interplay between the gut microbiota and host physiology in humans has been advanced by the human gut reference catalogue3,4. Thus, establishment of a comprehensive pig gut microbiome gene reference catalogue constitutes a logical continuation of the recently published pig genome5. By deep metagenome sequencing of faecal DNA from 287 pigs, we identified 7.7 million non-redundant genes representing 719 metagenomic species. Of the functional pathways found in the human catalogue, 96% are present in the pig catalogue, supporting the potential use of pigs for biomedical research. We show that sex, age and host genetics are likely to influence the pig gut microbiome. Analysis of the prevalence of antibiotic resistance genes demonstrated the effect of eliminating antibiotics from animal diets and thereby reducing the risk of spreading antibiotic resistance associated with farming systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lunney, J. K. Advances in swine biomedical model genomics. Int. J. Biol. Sci. 3, 179–184 (2007).
Sommer, F. & Bäckhed, F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Groenen, M. A. M. et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398 (2012).
Osei Sekyere, J. & Osei Sekyere, J. Antibiotic types and handling practices in disease management among pig farms in Ashanti region, Ghana. J. Vet. Med. 2014, e531952 (2014).
Aubry-Damon, H. et al. Antimicrobial resistance in commensal flora of pig farmers. Emerg. Infect. Dis. 10, 873–879 (2004).
Sayah, R. S., Kaneene, J. B., Johnson, Y. & Miller, R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal faecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71, 1394–1404 (2005).
Lammie, S. L. & Hughes, J. M. Antimicrobial resistance, food safety, and one health: the need for convergence. Annu. Rev. Food Sci. Technol. 7, 287–312 (2016).
Sanderson, H., Fricker, C., Brown, R. S., Majury, A. & Liss, S. N. Antibiotic resistance genes as an emerging environmental contaminant. Environ. Rev. 24 205–218 (2016).
Williams-Nguyen, J. et al. Antibiotics and antibiotic resistance in agroecosystems: state of the science. J. Environ. Qual. 45, 394–406 (2016).
Xiao, L. et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 33, 1103–1108 (2015).
Ai, H. et al. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics 15, 834 (2014).
Benson, A. K. et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA 107, 18933–18938 (2010).
Modi, S. R., Collins, J. J. & Relman, D. A. Antibiotics and the gut microbiota. J. Clin. Invest. 124, 4212–4218 (2014).
Looft, T. et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl Acad. Sci. USA 109, 1691–1696 (2012).
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A. & Collins, J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18 (2012).
Besemer, J., Lomsadze, A. & Borodovsky, M. Genemarks: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618 (2001).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Gerlach, W. & Stoye, J. Taxonomic classification of metagenomic shotgun sequences with CARMA3. Nucleic Acids Res. 39, e91 (2011).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Powell, S. et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 42, D231–D239 (2014).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2015).
Nielsen, H. B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828 (2014).
Dixon, P. & Palmer, M. W. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Bray, J. R. & Curtis, J. T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349 (1957).
Shepard, R. N. The analysis of proximities: multidimensional scaling with an unknown distance function. I. Psychometrika 27, 125–140 (1962).
Paulson, J. N., Stine, O. C., Bravo, H. C. & Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202 (2013).
Yamada, T., Letunic, I., Okuda, S., Kanehisa, M. & Bork, P. Ipath2.0: interactive pathway explorer. Nucleic Acids Res. 39, W412–W415 (2011).
Acknowledgements
This work was supported financially by the INRA metaprogramme Meta-omics of Microbial Ecosystems (MEM), the Animal Genetics Division of INRA, the Biology Department of the University of Copenhagen and BGI-Shenzhen. Y.R.C. was funded by the European Union, in the framework of the Marie-Curie FP7 COFUND People Program, through the award of an AgreenSkills' fellowship (grant agreement no. 267196) linked to the METALIT project funded by the INRA MEM metaprogramme. This study was supported by the Shenzhen Municipal Government of China (grant nos. JCYJ20140418095735538, DRC-SZ[2015]162, CXB201108250098A and CXZZ20150330171521403), the National Transgenic Project of China (2014ZX0801007B), the Science and Technology Plan of Shenzhen City, the Shenzhen–Hong Kong Innovation Circle Research Fund, China (SGLH20120928140607107). The work was also supported by the Danish Innovation Fund (grant nos. 4105-00011A and 0603-00774B) and Metagenopolis grant ANR-11-DPBS-0001. The authors acknowledge technical support for faecal stool sampling provided by the INRA experimental units (Y. Billon, UE GENESI at le Magneraud; C. Jaeger and H. Demay, UMR SENAH at Saint Gilles; N. Müller, UE0787 at le Rheu; E. Venturi and his team, UE PAO at Nouzilly; J.-L. Gourdine and D. Renaudeau, URZ in the Guadeloupe; J.-J. Leplat, UMR GABI at Jouy-en-Josas). The authors thank K. Pedersen at Svindinge farm, T. Povlsen at Blæsenborg farm, V. Jensen and K. Jensen at Stærsminde farm and H. Loft Hansen for technical assistance in relation to collection of faecal samples from the Danish farms. The authors also thank X. Liu and Zhigang at the Institute of Allergy & Immunology, Shenzhen University, and L. Fang Liang, Q. Wei and Y. Li at the technical unit of the Chinese farms for help with faecal sampling and administration. The authors thank C. Denis (INRA, UMR GABI, Jouy-en-Josas) and F. Levenez (INRA, UMR MICALIS and MetaGenoPolis, Jouy-en-Josas) for faecal DNA preparation. The authors acknowledge the @BRIDGe platform (INRA, Jouy-en-Josas) for safe storage of samples. The authors are grateful to J. Nathaniel Paulson for kind assistance with the use of the R package referred to as metagenomeSeq, and J. Lunney (Animal Parasitic Diseases Laboratory, ARS, USDA, MD, Beltsville, USA) for discussions and the contribution to manuscript editing.
Author information
Authors and Affiliations
Contributions
C.R.-G., J.W., L.M., E.M., J.D., S.D.E. and K.K. conceived and designed the project. K.K., C.R.-G., L.M., S.D.E., J.W., L.X., J.E., P.K., Y.R.-C., X.X., X.L., N.J.K. and Z.X. monitored the project. A.Ø.P., C.L. and E.P. collected samples and performed experiments. K.K., C.R.-G., L.M., J.W., J.E., Y.R.-C., P.K., N.P., E.L.C., E.P., S.D.E., Z.X., Q.F., S.L., A.Ø.P. and J.L. analysed and interpreted the data. K.K., C.R.-G., J.E., Y.R.-C., S.D.E., L.M., P.K., H.J. and L.X. wrote the paper. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1–8, legends for Supplementary Tables 1–9 (PDF 2038 kb)
Supplementary Table 1
Background information on the 287 pig samples. (XLSX 14 kb)
Supplementary Table 2
Description of the assembly data from the 287 samples. (XLSX 9 kb)
Supplementary Table 3
Assembly results of the pig, human and mouse data. (XLSX 10 kb)
Supplementary Table 4
Significantly differentially abundant genes in the gut microbiota composition between castrated males and females (Svindinge farm) at the genus and species bacteria levels. (XLSX 11 kb)
Supplementary Table 5
Significantly differentially represented KEGG pathways in the gut microbiota between castrated males and females (Svindinge farm). (XLSX 9 kb)
Supplementary Table 6
Significantly differentially abundant genes in the gut microbiota composition between males and females (Stæminde farm, wet feed) at the genus and species bacteria levels. (XLSX 12 kb)
Supplementary Table 7
Significantly differentially abundant MGSs in the gut microbiota composition of males and females (Stæminde farm, wet feed). (XLSX 17 kb)
Supplementary Table 8
Significantly differentially represented KEGG pathways in the gut microbiota between males and females (Stæminde farm, wet feed). (XLSX 54 kb)
Supplementary Table 9
List of the KEGG pathways mapped by iPATH2 that were significantly differentially represented in the gut microbiota between males and females (11 males, 14 females, Stæminde farm, wet feed). The three most represented pathways are highlighted in grey. This list is derived from Supplementary Table 8 and correspond to the differentially abundant functions mapping against the KEGG, according to iPATH2 tools. (XLSX 27 kb)
Rights and permissions
About this article
Cite this article
Xiao, L., Estellé, J., Kiilerich, P. et al. A reference gene catalogue of the pig gut microbiome. Nat Microbiol 1, 16161 (2016). https://doi.org/10.1038/nmicrobiol.2016.161
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.161
This article is cited by
-
Alternatives to antibiotics in pig production: looking through the lens of immunophysiology
Stress Biology (2024)
-
Effects of microbes in pig farms on occupational exposed persons and the environment
AMB Express (2023)
-
PandaGUT provides new insights into bacterial diversity, function, and resistome landscapes with implications for conservation
Microbiome (2023)
-
The effects of antibiotic use on the dynamics of the microbiome and resistome in pigs
Animal Microbiome (2023)
-
Longitudinal metagenomic study reveals the dynamics of fecal antibiotic resistome in pigs throughout the lifetime
Animal Microbiome (2023)