Abstract
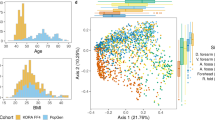
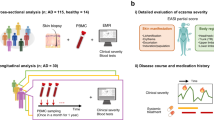
Whole metagenome analysis has the potential to reveal functional triggers of skin diseases, but issues of cost, robustness and sampling efficacy have limited its application. Here, we have established an alternative, clinically practical and robust metagenomic analysis protocol and applied it to 80 skin microbiome samples epidemiologically stratified for atopic dermatitis (AD). We have identified distinct non-flare, baseline skin microbiome signatures enriched for Streptococcus and Gemella but depleted for Dermacoccus in AD-prone versus normal healthy skin. Bacterial challenge assays using keratinocytes and monocyte-derived dendritic cells established distinct IL-1-mediated, innate and Th1-mediated adaptive immune responses with Staphylococcus aureus and Staphylococcus epidermidis. Bacterial differences were complemented by perturbations in the eukaryotic community and functional shifts in the microbiome-wide gene repertoire, which could exacerbate a dry and alkaline phenotype primed for pathogen growth and inflammation in AD-susceptible skin. These findings provide insights into how the skin microbial community, skin surface microenvironment and immune system cross-modulate each other, escalating the destructive feedback cycle between them that leads to AD flare.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Tay, Y. K., Kong, K. H., Khoo, L., Goh, C. L. & Giam, Y. C. The prevalence and descriptive epidemiology of atopic dermatitis in Singapore school children. Br. J. Dermatol. 146, 101–106 (2002).
Asher, M. I. et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet 368, 733–743 (2006).
Garmhausen, D. et al. Characterization of different courses of atopic dermatitis in adolescent and adult patients. Allergy 68, 498–506 (2013).
Paternoster, L. et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nature Genet. 47, 1449–1456 (2015).
Palmer, C. N. et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nature Genet. 38, 441–446 (2006).
van den Oord, R. A. & Sheikh, A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. Br. Med. J. 339, b2433 (2009).
Chen, H. et al. Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br. J. Dermatol. 165, 106–114 (2011).
Sandilands, A., Sutherland, C., Irvine, A. D. & McLean, W. H. Filaggrin in the frontline: role in skin barrier function and disease. J. Cell Sci. 122, 1285–1294 (2009).
Howell, M. D. et al. Th2 cytokines act on S100/A11 to downregulate keratinocyte differentiation. J. Invest. Dermatol. 128, 2248–2258 (2008).
Morizane, S. et al. TH2 cytokines increase kallikrein 7 expression and function in patients with atopic dermatitis. J. Allergy Clin. Immunol. 130, 259–61 e1 (2012).
Howell, M. D. et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J. Allergy Clin. Immunol. 124, R7–R12 (2009).
Hvid, M. et al. Regulation of caspase 14 expression in keratinocytes by inflammatory cytokines—a possible link between reduced skin barrier function and inflammation? Exp. Dermatol. 20, 633–636 (2011).
Brown, S. J. et al. Filaggrin haploinsufficiency is highly penetrant and is associated with increased severity of eczema: further delineation of the skin phenotype in a prospective epidemiological study of 792 school children. Br. J. Dermatol. 161, 884–889 (2009).
Fallon, P. G. et al. A homozygous frameshift mutation in the mouse Flg gene facilitates enhanced percutaneous allergen priming. Nature Genet. 41, 602–608 (2009).
Elias, P. M., Hatano, Y. & Williams, M. L. Basis for the barrier abnormality in atopic dermatitis: outside–inside–outside pathogenic mechanisms. J. Allergy Clin. Immunol. 121, 1337–1343 (2008).
Kabashima, K. New concept of the pathogenesis of atopic dermatitis: interplay among the barrier, allergy, and pruritus as a trinity. J. Dermatol. Sci. 70, 3–11 (2013).
Leyden, J. J., Marples, R. R. & Kligman, A. M. Staphylococcus aureus in the lesions of atopic dermatitis. Br. J. Dermatol. 90, 525–530 (1974).
Huang, J. T., Abrams, M., Tlougan, B., Rademaker, A. & Paller, A. S. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics 123, e808–e814 (2009).
Kong, H. H. et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012).
Oh, J. et al. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 23, 2103–2114 (2013).
Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014).
Andiappan, A. K. et al. Validation of GWAS loci for atopic dermatitis in a Singapore Chinese population. J. Invest. Dermatol. 132, 1505–1507 (2012).
Grice, E. A. et al. A diversity profile of the human skin microbiota. Genome Res. 18, 1043–1050 (2008).
Mathieu, A., Vogel, T. M. & Simonet, P. The future of skin metagenomics. Res. Microbiol. 165, 69–76 (2014).
Asher, M. I. et al. International study of asthma and allergies in childhood (ISAAC): rationale and methods. Eur. Respir. J. 8, 483–491 (1995).
Manivasagan, P., Venkatesan, J., Sivakumar, K. & Kim, S. K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 169, 262–278 (2014).
Chiller, K., Selkin, B. A. & Murakawa, G. J. Skin microflora and bacterial infections of the skin. J. Invest. Dermatol. Symp. Proc. 6, 170–174 (2001).
Wang, Y. et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 98, 411–424 (2014).
Schloissnig, S. et al. Genomic variation landscape of the human gut microbiome. Nature 493, 45–50 (2013).
Kapral, F. A., Smith, S. & Lal, D. The esterification of fatty acids by Staphylococcus aureus fatty acid modifying enzyme (FAME) and its inhibition by glycerides. J. Med. Microbiol. 37, 235–237 (1992).
Lowe, A. M., Beattie, D. T. & Deresiewicz, R. L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27, 967–976 (1998).
Mlckova, P., Cechova, D., Chalupna, P., Novotna, O. & Prokesova, L. Enhanced systemic and mucosal antibody responses to a model protein antigen after intranasal and intratracheal immunisation using Bacillus firmus as an adjuvant. Immunol. Lett. 77, 39–45 (2001).
Lutz, M. B. et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 223, 77–92 (1999).
Mitsui, H. et al. Differential expression and function of Toll-like receptors in Langerhans cells: comparison with splenic dendritic cells. J. Invest. Dermatol. 122, 95–102 (2004).
Malissen, B., Tamoutounour, S. & Henri, S. The origins and functions of dendritic cells and macrophages in the skin. Nature Rev. Immunol. 14, 417–428 (2014).
Lubbe, J. Secondary infections in patients with atopic dermatitis. Am. J. Clin. Dermatol. 4, 641–654 (2003).
Darabi, K., Hostetler, S. G., Bechtel, M. A. & Zirwas, M. The role of Malassezia in atopic dermatitis affecting the head and neck of adults. J. Am. Acad. Dermatol. 60, 125–136 (2009).
Wu, G. et al. Genus-wide comparative genomics of malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 11, e1005614 (2015).
Casagrande, B. F. et al. Sensitization to the yeast Malassezia sympodialis is specific for extrinsic and intrinsic atopic eczema. J. Invest. Dermatol. 126, 2414–2421 (2006).
Cork, M. J. et al. Epidermal barrier dysfunction in atopic dermatitis. J. Invest. Dermatol. 129, 1892–1908 (2009).
Moffett, J. R. & Namboodiri, M. A. Tryptophan and the immune response. Immunol. Cell Biol. 81, 247–265 (2003).
Lee, V. T. & Schneewind, O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15, 1725–1752 (2001).
Zgur-Bertok, D. DNA damage repair and bacterial pathogens. PLoS Pathogens 9, e1003711 (2013).
Cusumano, Z. T., Watson, M. E. Jr & Caparon, M. G. Streptococcus pyogenes arginine and citrulline catabolism promotes infection and modulates innate immunity. Infect. Immun. 82, 233–242 (2014).
Grice, E. A. & Segre, J. A. The skin microbiome. Nature Rev. Microbiol. 9, 244–253 (2011).
Dekio, I. et al. Characterization of skin microbiota in patients with atopic dermatitis and in normal subjects using 16S rRNA gene-based comprehensive analysis. J. Med. Microbiol. 56, 1675–1683 (2007).
Garcia-Garcera, M., Garcia-Etxebarria, K., Coscolla, M., Latorre, A. & Calafell, F. A new method for extracting skin microbes allows metagenomic analysis of whole-deep skin. PLoS One 8, e74914 (2013).
Oh, J., Conlan, S., Polley, E. C., Segre, J. A. & Kong, H. H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4, 77 (2012).
Conlan, S. et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13, R64 (2012).
Zhang, E. et al. Anti-malassezia-specific IgE antibodies production in Japanese patients with head and neck atopic dermatitis: relationship between the level of specific IgE antibody and the colonization frequency of cutaneous Malassezia species and clinical severity. J. Allergy (Cairo) 2011, 645670 (2011).
Andiappan, A. K. et al. Allergic airway diseases in a tropical urban environment are driven by dominant mono-specific sensitization against house dust mites. Allergy 69, 501–509 (2014).
Broccardo, C. J., Mahaffey, S. B., Strand, M., Reisdorph, N. A. & Leung, D. Y. Peeling off the layers: skin taping and a novel proteomics approach to study atopic dermatitis. J. Allergy Clin. Immunol. 124, 1113–1115 (2009).
Kezic, S., Kammeyer, A., Calkoen, F., Fluhr, J. W. & Bos, J. D. Natural moisturizing factor components in the stratum corneum as biomarkers of filaggrin genotype: evaluation of minimally invasive methods. Br. J. Dermatol. 161, 1098–1104 (2009).
Williamson, P. & Kligman, A. M. A new method for the quantitative investigation of cutaneous bacteria. J. Invest. Dermatol. 45, 498–503 (1965).
Pachtman, E. A., Vicher, E. E. & Brunner, M. J. The bacteriologic flora in seborrheic dermatitis. J. Invest. Dermatol. 22, 389–396 (1954).
Miller, C. S., Baker, B. J., Thomas, B. C., Singer, S. W. & Banfield, J. F. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 12, R44 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Ong, S. H. et al. Species identification and profiling of complex microbial communities using shotgun Illumina sequencing of 16S rRNA amplicon sequences. PLoS One 8, e60811 (2013).
Segata, N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods 9, 811–814 (2012).
Chen, L. et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328 (2005).
Wilm, A. et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40, 11189–11201 (2012).
Dickson, M. A. et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20, 1436–1447 (2000).
Malinarich, F. et al. High mitochondrial respiration and glycolytic capacity represent a metabolic phenotype of human tolerogenic dendritic cells. J. Immunol. 194, 5174–5186 (2015).
Haniffa, M. et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 37, 60–73 (2012).
Ginhoux, F. et al. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J. Exp. Med. 204, 3133–3146 (2007).
Hong, C. et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome 2, 33 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012).
Abubucker, S. et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358 (2012).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Acknowledgements
The authors thank all volunteers for donating their skin microbiome samples, the Biological Resource Centre at A*STAR for assistance with animal work, the Flow Cytometry Facility at the Singapore Immunology Network, A*STAR, for assistance with cell sorting, and G. Low, H. Chan and M. Unnaamalai for assistance with the isolation of epidermal LCs. This work was funded by an SRG National Healthcare Group grant to J.C., N.N. and M.B.Y.T. and A*STAR SPF funding for basic and translational skin research.
Author information
Authors and Affiliations
Contributions
K.R.C., M.B.Y.T., F.T.C., J.E.A.C. and N.N. planned the study. A.S.L.T., A.H.Q.N., J.W., N.M., B.J., X.F.C.C.W. and B.V.A. designed and conducted all experiments under the guidance of J.E.A.C., K.R.C., P.F.D.S., F.G. and J.E.C. Metagenomic data analysis was performed by K.R.C. and C.L. with supervision from N.N. Human skin tissue samples were collected by T.C.L. and A.W. provided technical computational assistance. B.K.S., S.A.M., Y.Y.S., F.T.C. and J.E.A.C. organized volunteer recruitment and sampling. K.R.C., A.S.L.T., E.B.L., J.E.A.C. and N.N. wrote the manuscript, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Notes 1–3, Supplementary References, Supplementary Figures 1–14, Supplementary Tables 1–3 and 8–10, Supplementary Tables 4–7 legends. (PDF 1224 kb)
Supplementary Table 4
Detailed metadata of study subjects. (XLSX 16 kb)
Supplementary Table 5
Table showing the relative abundances of different microbial genera and species in the skin microbiome of study subjects. (XLSX 99 kb)
Supplementary Table 6
Table showing pairwise Spearman correlations between the relative abundances of different skin microbial species across study subjects. (XLSX 17 kb)
Supplementary Table 7
Table showing SNPs of key staphylococcal virulence factors between cases and controls. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Chng, K., Tay, A., Li, C. et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat Microbiol 1, 16106 (2016). https://doi.org/10.1038/nmicrobiol.2016.106
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.106
This article is cited by
-
Novel methodologies for host-microbe interactions and microbiome-targeted therapeutics in 3D organotypic skin models
Microbiome (2023)
-
Skin microbiome differentiates into distinct cutotypes with unique metabolic functions upon exposure to polycyclic aromatic hydrocarbons
Microbiome (2023)
-
Microbiome in Atopic Dermatitis: Is It All About Staphylococcus aureus?
Current Treatment Options in Allergy (2023)
-
Controlling skin microbiome as a new bacteriotherapy for inflammatory skin diseases
Inflammation and Regeneration (2022)
-
Sarcoptic mange changes bacterial and fungal microbiota of bare-nosed wombats (Vombatus ursinus)
Parasites & Vectors (2022)