Abstract
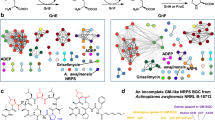
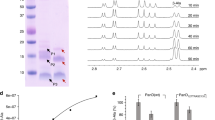
Phenotypic screens for bactericidal compounds against drug-resistant tuberculosis are beginning to yield novel inhibitors. However, reliable target identification remains challenging. Here, we show that tetrahydropyrazo[1,5-a]pyrimidine-3-carboxamide (THPP) selectively pulls down EchA6 in a stereospecific manner, instead of the previously assigned target Mycobacterium tuberculosis MmpL3. While homologous to mammalian enoyl-coenzyme A (CoA) hydratases, EchA6 is non-catalytic yet essential and binds long-chain acyl-CoAs. THPP inhibitors compete with CoA-binding, suppress mycolic acid synthesis, and are bactericidal in a mouse model of chronic tuberculosis infection. A point mutation, W133A, abrogated THPP-binding and increased both the in vitro minimum inhibitory concentration and the in vivo effective dose 99 in mice. Surprisingly, EchA6 interacts with selected enzymes of fatty acid synthase II (FAS-II) in bacterial two-hybrid assays, suggesting essentiality may be linked to feeding long-chain fatty acids to FAS-II. Finally, our data show that spontaneous resistance-conferring mutations can potentially obscure the actual target or alternative targets of small molecule inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
WHO. Global Tuberculosis Report 2013 (World Health Organization, 2013).
Abrahams, K. A. et al. Identification of novel imidazo[1,2-a]pyridine inhibitors targeting M. tuberculosis QcrB. PLoS ONE 7, e52951 (2012).
Gurcha, S. S. et al. Biochemical and structural characterization of mycobacterial aspartyl-tRNA synthetase AspS, a promising TB drug target. PLoS ONE 9, e113568 (2014).
Mugumbate, G. et al. Mycobacterial dihydrofolate reductase inhibitors identified using chemogenomic methods and in vitro validation. PLoS ONE 10, e0121492 (2015).
Wang, F. et al. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc. Natl Acad. Sci. USA 110, E2510–E2517 (2013).
Andries, K. et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227 (2005).
Grzegorzewicz, A. E. et al. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nature Chem. Biol. 8, 334–341 (2012).
La Rosa, V. et al. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob. Agents Chemother. 56, 324–331 (2012).
Li, K. et al. Multitarget drug discovery for tuberculosis and other infectious diseases. J. Med. Chem. 57, 3126–3139 (2014).
Li, W. et al. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 6413–6423 (2014).
Rao, S. P. et al. Indolcarboxamide is a preclinical candidate for treating multidrug-resistant tuberculosis. Sci. Transl. Med. 5, 214ra168 (2013).
Remuinan, M. J. et al. Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6′,7′-dihydrospiro[piperidine-4,4′-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PLoS ONE 8, e60933 (2013).
Tahlan, K. et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 1797–1809 (2012).
Banerjee, A. et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263, 227–230 (1994).
Zhang, Y., Heym, B., Allen, B., Young, D. & Cole, S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358, 591–593 (1992).
Bantscheff, M. et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nature Biotechnol. 29, 255–265 (2011).
Brown, A. K. et al. Identification of the dehydratase component of the mycobacterial mycolic acid-synthesizing fatty acid synthase-II complex. Microbiology 153, 4166–4173 (2007).
Rana, A. K. et al. Ppm1-encoded polyprenyl monophosphomannose synthase activity is essential for lipoglycan synthesis and survival in mycobacteria. PLoS ONE 7, e48211 (2012).
Kremer, L. et al. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J. Biol. Chem. 275, 16857–16864 (2000).
Cantaloube, S., Veyron-Churlet, R., Haddache, N., Daffe, M. & Zerbib, D. The Mycobacterium tuberculosis FAS-II dehydratases and methyltransferases define the specificity of the mycolic acid elongation complexes. PLoS ONE 6, e29564 (2011).
Bahnson, B. J., Anderson, V. E. & Petsko, G. A. Structural mechanism of enoyl-CoA hydratase: three atoms from a single water are added in either an E1cb stepwise or concerted fashion. Biochemistry 41, 2621–2629 (2002).
Rullas, J. et al. Fast standardized therapeutic-efficacy assay for drug discovery against tuberculosis. Antimicrob. Agents Chemother. 54, 2262–2264 (2010).
Engel, C. K., Mathieu, M., Zeelen, J. P., Hiltunen, J. K. & Wierenga, R. K. Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket. EMBO J. 15, 5135–5145 (1996).
Hamed, R. B., Batchelar, E. T., Clifton, I. J. & Schofield, C. J. Mechanisms and structures of crotonase superfamily enzymes—how nature controls enolate and oxyanion reactivity. Cell. Mol. Life Sci. 65, 2507–2527 (2008).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Hazbon, M. H. et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 50, 2640–2649 (2006).
Cole, S. T. et al. Massive gene decay in the leprosy bacillus. Nature 409, 1007–1011 (2001).
Sassetti, C. M., Boyd, D. H. & Rubin, E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 (2003).
Franceschini, A. et al. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013).
Kremer, L. et al. Mycolic acid biosynthesis and enzymic characterization of the beta-ketoacyl-ACP synthase A-condensing enzyme from Mycobacterium tuberculosis. Biochem. J. 364, 423–430 (2002).
Besra, G. S. et al. Identification of the apparent carrier in mycolic acid synthesis. Proc. Natl Acad. Sci. USA 91, 12735–12739 (1994).
Hu, Y. et al. 3-Ketosteroid 9α-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 75, 107–121 (2010).
Karimova, G., Pidoux, J., Ullmann, A. & Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl Acad. Sci. USA 95, 5752–5756 (1998).
Savitski, M. M. et al. Targeted data acquisition for improved reproducibility and robustness of proteomic mass spectrometry assays. J. Am. Soc. Mass Spectr. 21, 1668–1679 (2010).
Savitski, M. M. et al. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J. Proteome Res. 12, 3586–3598 (2013).
Savitski, M. M. et al. Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on Orbitrap-type mass spectrometers. Anal. Chem. 83, 8959–8967 (2011).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002).
Acknowledgements
G.S.B. acknowledges support in the form of a Personal Research Chair from J. Bardrick and a Royal Society Wolfson Research Merit Award. The research leading to these results has received funding from the European Union's 7th Framework Programme (FP7-2007-2013) under grant agreement no. 261378 and the BBSRC through an Industrial CASE studentship. The authors thank N. Zinn and T. Mathieson for mass spectrometry and database design, and S. Batt, P. Moynihan and L. Alderwick for technical support. The authors also acknowledge the TB Alliance for their discussions and expertise in the field.
Author information
Authors and Affiliations
Contributions
J.A.G.C., K.A.A., S.G.D., U.K., M.B., G.D., N.C.C., A.B., L.B., D.B., K.F. and G.S.B. conceived and designed the experiments. J.A.G.C., K.A.A., I.A.B., S.B., C.A., A.B., A.S., P.J.J., S.G.D., V.N., S.S.G., M.J.R., L.E. and J.R. performed the experiments. K.A.A., J.A.G.C., J.R., S.B., S.G.D., U.K., M.B., S.G.D., D.B., L.B., K.F. and G.S.B. analysed the data. L.B., M.J.P. and G.S.B. contributed reagents, materials and analysis tools. J.A.G.C., K.A.A., L.B., D.B., S.G.D., G.D., C.A., A.B., K.F. and G.S.B. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–6 and Tables 1–7 (PDF 19717 kb)
Rights and permissions
About this article
Cite this article
Cox, J., Abrahams, K., Alemparte, C. et al. THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria. Nat Microbiol 1, 15006 (2016). https://doi.org/10.1038/nmicrobiol.2015.6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2015.6
This article is cited by
-
The emerging role of mass spectrometry-based proteomics in drug discovery
Nature Reviews Drug Discovery (2022)
-
Identification and characterization of aspartyl-tRNA synthetase inhibitors against Mycobacterium tuberculosis by an integrated whole-cell target-based approach
Scientific Reports (2018)
-
Inhibiting mycobacterial tryptophan synthase by targeting the inter-subunit interface
Scientific Reports (2017)
-
Open Lab as a source of hits and leads against tuberculosis, malaria and kinetoplastid diseases
Nature Reviews Drug Discovery (2016)
-
Identification of KasA as the cellular target of an anti-tubercular scaffold
Nature Communications (2016)