Abstract
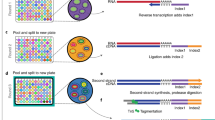
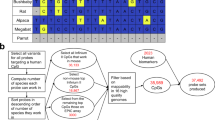
Microarrays—high-throughput platforms for analyzing the gene expression and features of total genomic DNA, among other applications—are gaining in popularity as researchers discover ever more uses for their unbiased and broad feature sets. At present, microarray analyses are limited by the number of individual features that can be placed on each array. Here we describe a double-tiling method that significantly increases the number of sequences present on an array, and we show that successful transcriptional profiling is possible and straightforward with such arrays. With this method, we and others can save money and precious samples by using fewer arrays to cover a region, or can carry out investigations at significantly higher resolution without incurring prohibitive costs or increasing the amount of sample required for the experiment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Bertone, P., Gerstein, M. & Snyder, M. Applications of DNA tiling arrays to experimental genome annotation and regulatory pathway discovery. Chromosome Res. 13, 259–274 (2005).
Bertone, P. et al. Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246 (2004).
Mockler, T.C. et al. Applications of DNA tiling arrays for whole-genome analysis. Genomics 85, 1–15 (2005).
Shoemaker, D.D. et al. Experimental annotation of the human genome using microarray technology. Nature 409, 922–927 (2001).
Lashkari, D.A. et al. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. USA 94, 13057–13062 (1997).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3 (2004).
Irizarry, R.A. et al. Multiple-laboratory comparison of microarray platforms. Nat. Methods 2, 345–350 (2005).
Cheng, J. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154 (2005).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000).
Wheelan, S.J., Scheifele, L.Z., Martínez-Murillo, F., Irizarry, R.A. & Boeke, J.D. The transposon insertion site profiling chip (JJP-chip). Proc. Natl. Acad. Sci. USA (in press).
Schmitt, M.E., Brown, T.A. & Trumpower, B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18, 3091–3092 (1990).
Acknowledgements
We thank H. Zhu and J. Bader for helpful comments on the manuscript and B. Greenlee for help with the figures and the methods section. Supported by grants from the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
S.J.W. designed arrays, executed one-color array experiments and prepared samples for two-color arrays, and analyzed data for one-color experiments; F.M.-M. executed two-color array experiments; R.A.I. analyzed data for two-color experiments; J.D.B. oversaw design, execution and analysis of all experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors have filed a patent application covering the technology described in this article.
Rights and permissions
About this article
Cite this article
Wheelan, S., Martínez-Murillo, F., Irizarry, R. et al. Stacking the deck: double-tiled DNA microarrays. Nat Methods 3, 903–907 (2006). https://doi.org/10.1038/nmeth951
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth951
This article is cited by
-
An efficient pseudomedian filter for tiling microrrays
BMC Bioinformatics (2007)