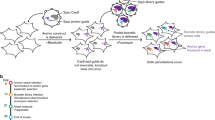
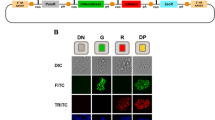
Abstract
RNA interference was originally described as a powerful tool to inhibit gene expression in model organisms. Until recently, loss-of-function genetic screens in mammalian cells were hampered by a lack of suitable tools that can be used in a high-throughput format. Here we discuss the construction of short-hairpin RNA (shRNA) vector libraries, in particular those generated at the Netherlands Cancer Institute (NKI), and their application in mammalian cancer genetics. We describe their virtues and limitations, as well as different options for screening such libraries.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Brummelkamp, T.R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J. & Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16, 948–958 (2002).
Brummelkamp, T.R., Bernards, R. & Agami, R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2, 243–247 (2002).
Dirac, A.M. & Bernards, R. Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J. Biol. Chem. 278, 11731–11734 (2003).
Michiels, F. et al. Arrayed adenoviral expression libraries for functional screening. Nat. Biotechnol. 20, 1154–1157 (2002).
Stegmeier, F., Hu, G., Rickles, R.J., Hannon, G.J. & Elledge, S.J. A lentiviral microRNA-based system for single-copy polymerase II–regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 102, 13212–13217 (2005).
Khvorova, A., Reynolds, A. & Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003).
Ngo, V.N. et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 (2006).
Van De Wetering, M. et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4, 609–615 (2003).
Berns, K. et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431–437 (2004).
Schwarz, D.S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Torrance, C.J., Agrawal, V., Vogelstein, B. & Kinzler, K.W. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat. Biotechnol. 19, 940–945 (2001).
Voorhoeve, P. & Agami, R. Tumor-suppressive functions of the human INK4A locus. Cancer Cell 4, 311–319 (2003).
Brummelkamp, T.R. & Bernards, R. New tools for functional mammalian cancer genetics. Nat. Rev. Cancer 3, 781–789 (2003).
Brummelkamp, T.R. et al. An shRNA bar code screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat. Chem. Biol. 2, 202–206 (2006).
Silva, J.M. et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 37, 1281–1288 (2005).
Paddison, P.J. et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 428, 427–431 (2004).
Brummelkamp, T.R. et al. Functional identification of cancer-relevant genes through large-scale RNA interference screens in mammalian cells. Cold Spring Harb. Symp. Quant. Biol. 69, 439–445 (2004).
Dirac, A.M., Nijman, S.M., Brummelkamp, T.R. & Bernards, R. Functional annotation of deubiquitinating enzymes using RNA interference. Methods Enzymol. 398, 554–567 (2005).
Westbrook, T.F. et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848 (2005).
Nijman, S.M. et al. The deubiquitinating enzyme USP1 regulates the Fanconi Anemia pathway. Mol. Cell 17, 331–339 (2005).
Kolfschoten, I.G. et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 121, 849–858 (2005).
Hartwell, L.H., Szankasi, P., Roberts, C.J., Murray, A.W. & Friend, S.H. Integrating genetic approaches into the discovery of anticancer drugs. Science 278, 1064–1068 (1997).
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006).
Brummelkamp, T.R., Nijman, S.M., Dirac, A.M. & Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797–801 (2003).
Nicke, B. et al. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol. Cell 20, 673–685 (2005).
Acknowledgements
We thank A. Fabius and A. Dirac for critical reading of the manuscript. This work was supported by grants from the Netherlands Genomics Initiative, The EU 6th framework integrated project “INTACT”, the centre for Biomedical genetics (CBG) and the Dutch Cancer Society (KWF).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
R.B. and R.L.B. receive royalties on sales of the shRNA library.
Rights and permissions
About this article
Cite this article
Bernards, R., Brummelkamp, T. & Beijersbergen, R. shRNA libraries and their use in cancer genetics. Nat Methods 3, 701–706 (2006). https://doi.org/10.1038/nmeth921
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth921
This article is cited by
-
Identification of acquired Notch3 dependency in metastatic Head and Neck Cancer
Communications Biology (2023)
-
Exploring liver cancer biology through functional genetic screens
Nature Reviews Gastroenterology & Hepatology (2021)
-
Kinase-targeted cancer therapies: progress, challenges and future directions
Molecular Cancer (2018)
-
Construction and Identification of the RNAi Recombinant Lentiviral Vector Targeting Human DEPDC7 Gene
Interdisciplinary Sciences: Computational Life Sciences (2017)
-
The utility of transposon mutagenesis for cancer studies in the era of genome editing
Genome Biology (2015)