Abstract
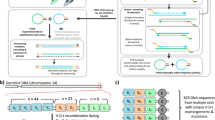
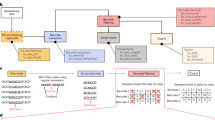
BEAMing allows the one-to-one conversion of a population of DNA fragments into a population of beads. We used rolling circle amplification to increase the number of copies bound to such beads by more than 100-fold. This allowed enumeration of mutant and wild-type sequences even when they were present at ratios less than 1:10,000 and was sensitive enough to directly quantify the error rate of DNA polymerases used for PCR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dressman, D., Yan, H., Traverso, G., Kinzler, K.W. & Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. USA 100, 8817–8822 (2003).
Diehl, F. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 102, 16368–16373 (2005).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Lizardi, P.M. et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19, 225–232 (1998).
Larsson, C. et al. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat. Methods 1, 227–232 (2004).
Tindall, K.R. & Kunkel, T.A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry 27, 6008–6013 (1988).
Bielas, J.H. & Loeb, L.A. Quantification of random genomic mutations. Nat. Methods 2, 285–290 (2005).
Li-Sucholeiki, X.C. & Thilly, W.G. A sensitive scanning technology for low frequency nuclear point mutations in human genomic DNA. Nucleic Acids Res. 28, E44 (2000).
Shi, C. et al. LigAmp for sensitive detection of single-nucleotide differences. Nat. Methods 1, 141–147 (2004).
Liu, Q. & Sommer, S.S. Detection of extremely rare alleles by bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification (Bi-PAP-A): measurement of mutation load in mammalian tissues. Biotechniques 36, 156–166 (2004).
Tawfik, D.S. & Griffiths, A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16, 652–656 (1998).
Acknowledgements
We thank L. Meszler for the help with imaging, Y. He and S. Zhou for advice. This work was supported by the US National Colorectal Cancer Research Alliance, The Clayton Fund and National Institutes of Health grants CA43460, CA57345 and CA62924.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Under a licensing agreement between EXACT Sciences and The Johns Hopkins University, K.W.K. and B.V. are entitled to a share of royalties received by the university on sales of products related to digital PCR. Under a licensing agreement between Agencourt Biosciences Corporation and The Johns Hopkins University, D.D., K.W.K. and B.V. are entitled to a share of royalties received by the university on sales of products related to the use of BEAMing for preparing templates for DNA sequencing. The terms of these arrangements are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
Supplementary information
Supplementary Fig. 1
Relationship between template DNA and single-template beads produced by BEAMing. (PDF 419 kb)
Supplementary Fig. 2
Relationship between input template ratio and bead proportions generated by BEAMing. (PDF 395 kb)
Supplementary Fig. 3
Signal to noise ratio of rolling circle amplification (RCA) on beads. (PDF 64 kb)
Supplementary Fig. 4
Quantification of beads produced by BEAMing Up using templates representing mixtures of wt and mutant TP53 sequences. (PDF 37 kb)
Supplementary Fig. 5
Quantification of beads produced by BEAMing Up using templates representing mixtures of wt and mutant PIK3CA sequences. (PDF 489 kb)
Supplementary Fig. 6
Quantification of error rates of representative DNA polymerases used for PCR. (PDF 444 kb)
Supplementary Table 1
Statistical examples of means and standard deviations of flow cytometric profiles for templates containing 0.01% TP53 mutant sequences. (PDF 24 kb)
Rights and permissions
About this article
Cite this article
Li, M., Diehl, F., Dressman, D. et al. BEAMing up for detection and quantification of rare sequence variants. Nat Methods 3, 95–97 (2006). https://doi.org/10.1038/nmeth850
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth850
This article is cited by
-
Current status of molecular diagnostic approaches using liquid biopsy
Journal of Gastroenterology (2023)
-
Advances in optical counting and imaging of micro/nano single-entity reactors for biomolecular analysis
Analytical and Bioanalytical Chemistry (2023)
-
Sensitive detection of low-abundance in-frame deletions in EGFR exon 19 using novel wild-type blockers in real-time PCR
Scientific Reports (2019)
-
Progress toward liquid biopsies in pediatric solid tumors
Cancer and Metastasis Reviews (2019)
-
Utility of targeted deep sequencing for detecting circulating tumor DNA in pancreatic cancer patients
Scientific Reports (2018)