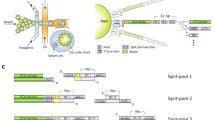
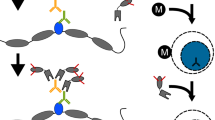
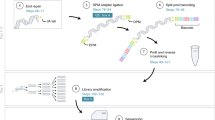
Abstract
High-throughput gene analysis would benefit from new approaches for delivering DNA or RNA into cells. Here we describe a simple system that allows any molecular biology laboratory to carry out multiple, parallel cell transfections on microscope coverslip arrays. By using magnetically defined positions and PCR product–coated paramagnetic beads, we achieved transfection in a variety of cell lines. Beads may be added to the cells at any time, allowing both spatial and temporal control of transfection. Because the beads may be coated with more than one gene construct, the method can be used to achieve cotransfection within single cells. Furthermore, PCR-generated mutants may be conveniently screened, bypassing cloning and plasmid purification steps. We illustrated the applicability of the method by screening combinatorial peptide libraries, fused to GFP, to identify previously unknown cellular localization motifs. In this way, we identified several localizing peptides, including structured localization signals based around the scaffold of a single C2H2 zinc finger.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ziauddin, J. & Sabatini, D.M. Microarrays of cells expressing defined cDNAs. Nature 411, 107–110 (2001).
Erfle, H., Simpson, J.C., Bastiaens, P.I. & Pepperkok, R. siRNA cell arrays for high-content screening microscopy. Biotechniques 37, 454–458, 460, 462 (2004).
Bejarano, L.A. & Gonzalez, C. Motif trap: a rapid method to clone motifs that can target proteins to defined subcellular localisations. J. Cell Sci. 112, 4207–4211 (1999).
Kamath, R.S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003).
Uetz, P. et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000).
Wu, R.Z., Bailey, S.N. & Sabatini, D.M. Cell-biological applications of transfected-cell microarrays. Trends Cell Biol. 12, 485–488 (2002).
Hasty, J., McMillen, D. & Collins, J.J. Engineered gene circuits. Nature 420, 224–230 (2002).
Guet, C.C., Elowitz, M.B., Hsing, W. & Leibler, S. Combinatorial synthesis of genetic networks. Science 296, 1466–1470 (2002).
Cormack, B.P., Valdivia, R.H. & Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 (1996).
Gurskaya, N.G. et al. GFP-like chromoproteins as a source of far-red fluorescent proteins. FEBS Lett. 507, 16–20 (2001).
Chen, F., MacDonald, C.C. & Wilusz, J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 23, 2614–2620 (1995).
Luo, Y., Batalao, A., Zhou, H. & Zhu, L. Mammalian two-hybrid system: a complementary approach to the yeast two-hybrid system. Biotechniques 22, 350–352 (1997).
Kalderon, D., Roberts, B.L., Richardson, W.D. & Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 39, 499–509 (1984).
Isalan, M., Klug, A. & Choo, Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 19, 656–660 (2001).
Gould, S.G., Keller, G.A. & Subramani, S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J. Cell Biol. 105, 2923–2931 (1987).
Nair, R., Carter, P. & Rost, B. NLSdb: database of nuclear localization signals. Nucleic Acids Res. 31, 397–399 (2003).
Munro, S. & Pelham, H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 (1987).
Recalcati, S., Menotti, E. & Kuhn, L.C. Peroxisomal targeting of mammalian hydroxyacid oxidase 1 requires the C-terminal tripeptide SKI. J. Cell Sci. 114, 1625–1629 (2001).
Emanuelsson, O., Nielsen, H., Brunak, S. & von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 (2000).
Lacroix, E., Viguera, A.R. & Serrano, L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 284, 173–191 (1998).
Pearson, W.R. & Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85, 2444–2448 (1988).
Palenik, B. et al. The genome of a motile marine Synechococcus. Nature 424, 1037–1042 (2003).
Pavletich, N.P. & Pabo, C.O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science 252, 809–817 (1991).
Mackay, J.P. & Crossley, M. Zinc fingers are sticking together. Trends Biochem. Sci. 23, 1–4 (1998).
Isalan, M. Zinc fingers. in Encyclopedia of Biological Chemistry (eds. Lennars, W.J. & Lane, M.D.) (Academic Press–Elsevier, St. Louis, 2004).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Bailey, S.N., Wu, R.Z. & Sabatini, D.M. Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discov. Today 7, S113–S118 (2002).
Grimm, S. & Kachel, V. Robotic high-throughput assay for isolating apoptosis-inducing genes. Biotechniques 32, 670–2, 674–7 (2002).
Liebel, U. et al. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 554, 394–398 (2003).
Mousses, S. et al. RNAi microarray analysis in cultured mammalian cells. Genome Res. 13, 2341–2347 (2003).
Silva, J.M., Mizuno, H., Brady, A., Lucito, R. & Hannon, G.J. RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc. Natl. Acad. Sci. USA 101, 6548–6552 (2004).
Acknowledgements
M.I. was supported by an International Research Fellowship from the Wellcome Trust, UK. M.I.S. was funded by a Fundacion Carolina Postdoctoral Fellowship. We would like to thank G. DeCarcer Diez and K. Michalodimitrakis for help in the design of the transfection chamber and D. Megias Vazquez for assistance with confocal imaging.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Isalan, M., Santori, M., Gonzalez, C. et al. Localized transfection on arrays of magnetic beads coated with PCR products. Nat Methods 2, 113–118 (2005). https://doi.org/10.1038/nmeth732
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth732
This article is cited by
-
The living microarray: a high-throughput platform for measuring transcription dynamics in single cells
BMC Genomics (2011)
-
Cloning-free regulated monitoring of reporter and gene expression
BMC Molecular Biology (2009)
-
Evolvability and hierarchy in rewired bacterial gene networks
Nature (2008)
-
Generation of magnetic nonviral gene transfer agents and magnetofection in vitro
Nature Protocols (2007)
-
Construction of semi-randomized gene libraries with weighted oligonucleotide synthesis and PCR
Nature Protocols (2006)