Abstract
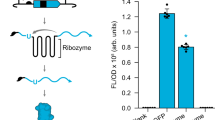
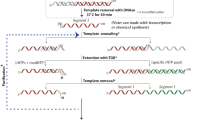
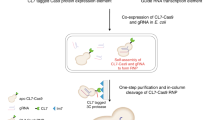
RNA has emerged as a major player in most cellular processes. Understanding these processes at the molecular level requires homogeneous RNA samples for structural, biochemical and pharmacological studies. So far, this has been a bottleneck, as the only methods for producing such pure RNA have been in vitro syntheses. Here we describe a generic approach for expressing and purifying structured RNA in Escherichia coli, using tools that parallel those available for recombinant proteins. Our system is based on a camouflage strategy, the 'tRNA scaffold', in which the recombinant RNA is disguised as a natural RNA and thus hijacks the host machinery, escaping cellular RNases. This opens the way to large-scale structural and molecular investigations of RNA function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Breaker, R.R. Natural and engineered nucleic acids as tools to explore biology. Nature 432, 838–845 (2004).
Holbrook, S.R. RNA structure: the long and the short of it. Curr. Opin. Struct. Biol. 15, 302–308 (2005).
Puerta-Fernandez, E., Romero-Lopez, C., Barroso-del Jesus, A. & Berzal-Herranz, A. Ribozymes: recent advances in the development of RNA tools. FEMS Microbiol. Rev. 27, 75–97 (2003).
Hermann, T. & Westhof, E. RNA as a drug target: chemical, modelling, and evolutionary tools. Curr. Opin. Biotechnol. 9, 66–73 (1998).
Jaeger, L. & Chworos, A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 16, 531–543 (2006).
Guo, P. RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy. J. Nanosci. Nanotechnol. 5, 1964–1982 (2005).
Milligan, J.F., Groebe, D.R., Witherell, G.W. & Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 (1987).
Marshall, W.S. & Kaiser, R.J. Recent advances in the high-speed solid phase synthesis of RNA. Curr. Opin. Chem. Biol. 8, 222–229 (2004).
Studier, F.W., Rosenberg, A.H., Dunn, J.J. & Dubendorff, J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60–89 (1990).
Moore, P.B. et al. Preparation of 5S RNA-related materials for nuclear magnetic resonance and crystallography studies. Methods Enzymol. 164, 158–174 (1988).
Masson, J.-M. & Miller, J.H. Expression of synthetic tRNA genes under the control of a synthetic promoter. Gene 47, 179–183 (1986).
Meinnel, T., Mechulam, Y. & Fayat, G. Fast purification of a functional elongator tRNAmet expressed from a synthetic gene in vivo. Nucleic Acids Res. 16, 8095–8096 (1988).
Tisné, C., Rigourd, M., Marquet, R., Ehresmann, C. & Dardel, F. NMR and biochemical characterization of recombinant human tRNA(Lys)3 expressed in Escherichia coli: identification of posttranscriptional nucleotide modifications required for efficient initiation of HIV-1 reverse transcription. RNA 6, 1403–1412 (2000).
Wallis, N.G., Dardel, F. & Blanquet, S. Heteronuclear NMR studies of the interactions of 15N-labeled methionine-specific transfer RNAs with methionyl-tRNA transformylase. Biochemistry 34, 7668–7677 (1995).
Deutscher, M.P. Ribonucleases, tRNA nucleotidyltransferase, and the 3′ processing of tRNA. Prog. Nucleic Acid Res. Mol. Biol. 39, 209–240 (1990).
Keiler, K.C., Waller, P.R. & Sauer, R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993 (1996).
Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K. & Inokuchi, H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli . Proc. Natl. Acad. Sci. USA 91, 9223–9227 (1994).
Junker-Niepmann, M., Bartenschlager, R. & Schaller, H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9, 3389–3396 (1990).
Nassal, M. & Rieger, A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70, 2764–2773 (1996).
Budker, V.G. et al. Photoaffinity reagents for modification of aminoacyl-tRNA synthetases. FEBS Lett. 49, 159–162 (1974).
Srisawat, C., Goldstein, I.J. & Engelke, D.R. Sephadex-binding RNA ligands: rapid affinity purification of RNA from complex RNA mixtures. Nucleic Acids Res. 29, E4 (2001).
Srisawat, C. & Engelke, D.R. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA 7, 632–641 (2001).
Grate, D. & Wilson, C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc. Natl. Acad. Sci. USA 96, 6131–6136 (1999).
Paillart, J.C., Skripkin, E., Ehresmann, B., Ehresmann, C. & Marquet, R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 93, 5572–5577 (1996).
Guo, P., Zhang, C., Chen, C., Garver, K. & Trottier, M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell 2, 149–155 (1998).
Zhang, F. et al. Function of hexameric RNA in packaging of bacteriophage phi 29 DNA in vitro. Mol. Cell 2, 141–147 (1998).
Raibaud, S. et al. How bacterial ribosomal protein L20 assembles with 23S ribosomal RNA and its own messenger RNA. J. Biol. Chem. 278, 36522–36530 (2003).
Price, S.R., Ito, N., Oubridge, C., Avis, J.M. & Nagai, K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 249, 398–408 (1995).
Lapham, J. & Crothers, D.M. RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA 2, 289–296 (1996).
Tisné, C. & Dardel, F. Optimisation of a peptide library for screening specific RNA ligands by flow-injection NMR. Comb. Chem. High Throughput Screen. 5, 523–529 (2002).
Tisné, C., Roques, B.P. & Dardel, F. The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J. Biol. Chem. 279, 3588–3595 (2004).
Acknowledgements
This research was supported by the 6th framework program of the European Union (grant FSG-V-RNA). We thank F. Allemand (Institut de Biologie Physico-Chimique, Paris) for the kind gift of purified E. coli L20 protein and C. Tisné and S. Nonin for assistance with the NMR experiments.
Author information
Authors and Affiliations
Contributions
F.D. was responsible for project planning. L.P. performed the experiments. F.D. and L.P. discussed the results, drafted the paper and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures and Text
Supplementary Figures 1-5, Supplementary Table 1. (PDF 1921 kb)
Rights and permissions
About this article
Cite this article
Ponchon, L., Dardel, F. Recombinant RNA technology: the tRNA scaffold. Nat Methods 4, 571–576 (2007). https://doi.org/10.1038/nmeth1058
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1058
This article is cited by
-
DNA scaffold assisted ectoine production in Escherichia coli
Systems Microbiology and Biomanufacturing (2024)
-
Avidity-based bright and photostable light-up aptamers for single-molecule mRNA imaging
Nature Chemical Biology (2023)
-
Structures of flavivirus RNA promoters suggest two binding modes with NS5 polymerase
Nature Communications (2021)