Abstract
The throughput of cell mechanical characterization has recently approached that of conventional flow cytometers. However, this very sensitive, label-free approach still lacks the specificity of molecular markers. Here we developed an approach that combines real-time 1D-imaging fluorescence and deformability cytometry in one instrument (RT-FDC), thus opening many new research avenues. We demonstrated its utility by using subcellular fluorescence localization to identify mitotic cells and test for mechanical changes in those cells in an RNA interference screen.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Shapiro, M.H. Practical Flow Cytometry (John Wiley & Sons, Inc., 2003).
Elson, E.L. Annu. Rev. Biophys. Biophys. Chem. 17, 397–430 (1988).
Fletcher, D.A. & Mullins, R.D. Nature 463, 485–492 (2010).
Matthews, H.K. et al. Dev. Cell 23, 371–383 (2012).
Otto, O. et al. Nat. Methods 12, 199–202 (2015).
Ekpenyong, A.E. et al. PLoS One 7, e45237 (2012).
Tse, H.T.K. et al. Sci. Transl. Med. 5, 212ra163 (2013).
Guck, J. et al. Biophys. J. 88, 3689–3698 (2005).
Worthen, G.S., Schwab, B.I., Elson, E.L. & Downey, G.P. Science 245, 183–186 (1989).
Bufi, N. et al. Biophys. J. 108, 2181–2190 (2015).
Toepfner, N. et al. eLife 7, e29213 (2018).
Gossett, D.R. et al. Proc. Natl. Acad. Sci. USA 109, 7630–7635 (2012).
Nyberg, K.D. et al. Biophys. J. 113, 1574–1584 (2017).
Ramdzan, Y.M. et al. Nat. Methods 9, 467–470 (2012).
Gray, J.W., Peters, D., Merrill, J.T., Martin, R. & Van Dilla, M.A. J. Histochem. Cytochem. 27, 441–444 (1979).
Basiji, D.A., Ortyn, W.E., Liang, L., Venkatachalam, V. & Morrissey, P. Clin. Lab. Med. 27, 653–670 (2007).
Lau, A.K.S., Shum, H.C., Wong, K.K. & Tsia, K.K. Lab Chip 16, 1743–1756 (2016).
Mietke, A. et al. Biophys. J. 109, 2023–2036 (2015).
Mokbel, M. et al. ACS Biomater. Sci. Eng. 3, 2962–2973 (2017).
Kräter, M. et al. Sci. Rep. 7, 2549 (2017).
Suzuki, S. Comput. Vis. Graph. Image Process. 46, 32–46 (1985).
Baum, B. & Cherbas, L. Methods Mol. Biol. 420, 391–424 (2008).
Herbig, M. et al. in Flow Cytometry Protocols (eds. Hawley, R. & Hawley, T.) 347–370 (Humana Press, 2017).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2015).
Bates, D. R News 5, 27–30 (2005).
Mood, A.M., Graybill, F.A. & Boes, D.C. Introduction to the Theory of Statistics 3rd ed. 540–541 (McGraw-Hill, 1974).
Wilks, S.S. Annals of Mathematical Statistics 9, 60–62 (1938).
Acknowledgements
We thank D. Klaue, M. Urbanska, F. Rosendahl, and I. Richter for helpful discussions and technical support. We thank M. Schürmann for refractive index measurements of agarose. We thank M. Piel (Institute Curie, Paris, France) for donation of the HeLa H2B–mCherry cell line. We thank M. Bornhäuser (Department of Hematology and Oncology, University Clinic Carl Gustav Carus, Technische Universität Dresden, Dresden, Germany) for providing the apheresis blood samples. We received support from the Light Microscopy Facility for confocal microscopy, and from the Microstructure Facility for microfluidic chip production; both are core facilities of the Center for Molecular and Cellular Bioengineering (CMCB) at Technische Universität Dresden. We gratefully acknowledge financial support from the Alexander-von-Humboldt Stiftung (Humboldt Professorship to J.G.), Sächsisches Ministerium für Wissenschaft und Kunst (TG70 grant to O.O. and J.G.; European Fund for Regional Development–EFRE to S.G. and J.G.), the ERC (starting grant “LightTouch” 282060 to J.G.), the DFG Center for Regenerative Medicine of the Technische Universität Dresden (seed grant to J.G.), the DFG KFO249 (GU 612/2-2 grant to J.G.), DKMS ('Mechthild Harf Research Grant' DKMS-SLS-MHG-2016-02 to A.J.), and Cancer Research UK (C1529/A17343 grant to B.B.).
Author information
Authors and Affiliations
Contributions
J.G. conceived the project. P.R. built the experimental setup. O.O. and C.H. supported the optics design. P.R. programmed the real-time data-evaluation software. K.P. designed and performed experiments on Kc167 and HeLa cells and analyzed the data with B.B.'s support. P.R. and N.T. performed experiments with RBCs. A.J., M.K., and P.R. performed experiments on HSPCs and analyzed the data. P.R. performed and analyzed experiments for device characterization. M.H., O.O., and Y.G. developed the linear mixed model algorithm for statistical evaluation. K.W. and S.G. produced fluorescent beads for device characterization. M.W. performed reference measurements with calibration beads on FCM. S.G. produced masters for microfluidic chips. J.G. and B.B. supervised the project. P.R., K.P., and J.G. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.R., O.O., and C.H. are shareholders and employees of the company Zellmechanik Dresden GmbH, which sells devices based on RT-DC and RT-FDC technology. All other authors declare no competing interests.
Integrated supplementary information
Supplementary Figure 1 Microfluidic chip layout.
Positions where inlet and outlet tubing are connected are marked with ⊗. Arrays of pillars filter out debris and dust particles to avoid clogging of the narrow channel. The region of interest of the camera is positioned at the end of the 300 μm long channel during the measurement (measurement region).
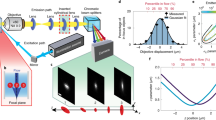
Supplementary Figure 2 Principle and characterization of the fluorescence detection of RT-FDC.
(a) Light sheet excitation and detection of fluorescence results in fluorescence peaks with widths depending on fluorophore localization. (b) Aperture function in focal plane (x-y) of excitation and detection for the three fluorescence channels/lasers. (c) Determination of thickness of the light sheet along the channel axis x yielded values of 3.64 ± 0.03 μm for 488 nm, 3.16 ± 0.02 μm for 561 nm, and 3.41 ± 0.05 μm for 640 nm. Thus, the line width is smaller than most eukaryotic cells and it can be used to identify sub-cellular distribution of fluorescent probes. (d) Measurement of two-color fluorescent agarose beads with heterogeneous sizes. Plotted are the fluorescence peak widths of the green (FL-1: 525/50 nm) and red (FL-3: 700/75 nm) channels. The linear fit indicates that the system determines nearly identical sizes in both channels. Experiments a-c were repeated three times with similar results.
Supplementary Figure 3 Calibration bead measurements.
Calibration beads with 8 populations were measured with RT-FDC and FCM (BD LSR II) for all three channels. (a-c) Fluorescence intensity histograms from RT-FDC measurements. Small labels in graph denote the peak position. (a) Green FL-1 channel (525/50 nm), (b) red FL-2 channel (593/46 nm), and (c) deep red FL-3 channel (700/75 nm). (d) Normalized sub-population intensities are compared versus peak or population index. (e-g) Histograms from LSR II FCM data for corresponding channels. (e) Green channel (530/30 nm), (f) red channel (575/26 nm), and (g) deep red channel (670/14 nm). Both methods consistently identify eight populations of objects with intensities over a dynamic range of three orders of magnitude. Experiments a-b, e-g were repeated three times with similar results.
Supplementary Figure 4 Evaluation of CD34+ subpopulation regarding image-processing-based parameters deformation, size, brightness, and granularity.
CD34− cells are plotted in gray while CD34+ cells are shown in colors that encode density of data points. (a) Deformation of CD34+ HSPCs vs. CD34-APC fluorescence intensity. (b) Projected area vs. CD34-APC fluorescence intensity. (c-d) Image-based brightness evaluation. (c) Brightness inside cell contour plotted over the CD34-APC intensity. Brightness can be used as an additional marker to identify subsets of leukocytes, as recently shown in Toepfner et al.11. (d) Granularity is characterized by the standard deviation of the pixel brightness values inside the cell contour, and plotted over CD34-APC intensity. There was no correlation of CD34-expression level detectable with either parameter. However, the CD34-positive HSPCs seem to have relatively low spread in the brightness parameters compared to the very heterogeneous CD34-negative cells. Experiments (a-d) were repeated three times independently with similar results.
Supplementary Figure 5 Three-color antibody staining of HSPCs.
Blood samples from GSF mobilized donors were collected after apheresis and stained with CD3-FITC, CD34-PE, and CD14-APC. (a) Gating strategy for exclusion of RBCs by cell size and brightness. (b) Gate for CD34-positive cells by fluorescence intensity. (c) Gate for CD3−CD14− subpopulation for exclusion by lineage markers. (d) Resulting mechanical fingerprint for CD34+CD3−CD14− subpopulation. (e) Back-gated subpopulation from (d) in initial representation. (f) CD3+ fraction in mechanical fingerprint. (g) CD14+ fraction. (h) CD34+ fraction with all other cells shown in gray. Experiment was performed once.
Supplementary Figure 6 Representative flow cytometry measurement of CD34–APC-stained G-CSF-mobilized blood samples.
(a) Forward scatter vs. side scatter plot with gate 1 to separate cell events from debris. (b) Fluorescently labeled CD34+ cells are found in gate 2 in the side scatter vs. APC intensity plot. (c) Histogram of fluorescence peak heights for cells of gate 1 (gate 3 = CD34+). Experiment was performed three times independently with similar results.
Supplementary Figure 7 The mechanical properties of immature versus mature red blood cells (RBCs).
Reticulocytes are the immature fraction of RBCs and represent normally about 5 – 15 ‰ of the total number of RBCs in circulating human blood12. During the first two days after being released to the blood stream, reticulocytes lose their cytoplasmic RNA as they mature to RBCs. This enabled us to use nucleic acid staining with syto13 to identify RNA-positive reticulocytes by fluorescence. Fluorescence read-out is compared between commercial LSR II FCM (a) and RT-FDC (b) as shown in the histograms. (c) Comparison of reticulocyte fraction determined with RT-FDC and state-of-the-art automatic blood count (XE-5000, Sysmex Deutschland) for three donors. Columns show counts, error bars represent standard error resulting from Poisson statistics. Reticulocyte numbers relative to RBCs were in agreement. (d) Location of RBC and reticulocyte population medians in deformation vs. area plot. (Shaded regions are for illustration purposes) Red blood cells have median size of 35.90 μm2 and deformation of 0.254 while reticulocytes are larger (39.2 μm2) and show smaller deformation (0.240). The difference indicated by arrows was significant as tested with linear mixed model analysis (Supplementary Table 2). (e,f) Representative deformation vs. area plots of reticulocytes and mature RBCs. (g) Deformation vs. syto13 fluorescence, and (h) area vs. syto13 fluorescence. Results are in line with previous reports on reticulocytes13, which had been restricted to labor-intensive micropipette and micro-pore assays with limited number of cells. We found human reticulocytes in whole blood samples to be slightly larger and less deformed compared to RBCs (d, Supplementary Table 2). Increased reticulocytosis has been linked with increased morbidity in infants with sickle cell anemia14 and RBC distribution width is also used as a predictor of poor clinical outcome in settings of various diseases including coronary heart disease15. Adding RT-FDC phenotyping to the reticulocyte indices in clinical use could therefore provide a means of monitoring changes of blood rheology in haematological diseases11,15. Experiments (a,b) were performed once, (c-h) were performed with samples from three different donors.
Supplementary Figure 8 1D-fluorophore localization detection by fluorescence peak width.
We used 1D imaging to detect the subcellular, nuclear localization of a fluorescent protein by comparing fluorescence peak widths for cells expressing nuclear localization sequence (nls) tagged and cytoplasmic (cyt) fluorescent proteins. Here, HeLa cells were transfected with plasmids expressing GFP and mRFP with and without nuclear locator sequence (nls). Red dots: cytosolic RFP and nuclear GFP, black dots: both fluorophores cytosolic, green dots cytosolic GFP and nuclear RFP. Cells where GFP and RFP fluorophores co-localize can be found on the diagonal of this plot. Since, in animal cells, cell cycle progression involves changes in nuclear permeability, we were able to exploit this feature to identify mitotic cells in heterogeneous population to identify mitotic cells in the Drosophila RNAi screen (see Fig. 2c). This experiment also serves to show that the green and red channel can be used interchangeably for this kind of analysis. Experiments were performed 3 times independently with similar results.
Supplementary Figure 9 Effect of colchicine treatment on fraction of mitotic Kc167 cells.
Fluorescence peak width plots (tdTomato vs. nls-GFP) for (a) non-treated or (b) colchicine treated cells show that colchicine treatment increases the number of cells with cytosolic nls-GFP (fraction of cells with even peak widths, localized along diagonal). During mitotic entry, the nuclear envelope breaks down releasing the nls-GFP protein to the cytosol, which affects the measured peak width in the GFP channel. This is exploited to identify mitotic cells in RT-FDC measurements. Mitotic cells represented by colored dots, interphase cells shown in gray. (c) Comparison with manually inspected confocal microscopy images of Hoechst33342 stained cells. Bar graphs show percentages obtained from counting, error bars indicate standard error resulting from Poisson statistics. For microcopy non-sync. 49/1063 (mitotic/total) cells were analyzed, for microscopy sync. 156/928 cells, and for RT-FDC non-sync. 388/6871 and 1078/6120 cells were analyzed. Experiments (a,b,c) were performed once.
Supplementary Figure 10 Data processing of Kc167 RNAi screen.
For each gene, experiments with two dsRNA variants were performed, each repeated on three different days and two different flow-rates. Exemplary graphs for gene 10 (Rok) at 0.04 μL/s flow-rate are shown. The statistical analysis for this and all other sets of measurements is available in Supplementary Table 3. Mitotic cells are represented by colored dots while limited number of interphase cells is shown in gray. All experiments (center column) were compared against a mock control (left column) from the same day. See right column for comparison of the scatter plots. Dashed contour lines are drawn at 50 % of the maximum density of data points, and solid contour lines at 95 %. Data from mock and treatment were transformed into elastic modulus values for comparison by the linear mixed model analysis (Supplementary Fig. 10 and Supplementary Table 3).
Supplementary Figure 11 Results of the screen for genes regulating mitotic cell mechanics in Kc167 cells.
For each gene two dsRNA constructs were used and each dsRNA treatment was repeated three times on separate days. The size and deformation data was transformed into E modulus using Shapeout software and statistical analysis was performed using a linear mixed model. Genes with consistent results for both flowrates and both p-values <0.05 were considered as hits. (“*”: p<0.05, “**”: p<0.01)
Supplementary Figure 12 Two-color fluorescence detection of FUCCI cell-cycle marker16.
(a) Two color fluorescence scatter plot with gates for G1, S and G2/M phase. (b) A pulse width evaluation enables detection of mitotic cells because of GFP localization inside the nucleus during interphase and in the cytosol during mitosis, resulting in wider pulses. (c-f) Cell cycle specific mechanical fingerprints for the mixed sample for G1, S, G2 and mitosis. Experiments were performed 10 times independently with similar results.
Supplementary Figure 13 Effect of suspension on RPE1 cells.
(a) Cell cycle phase-specific mechanical fingerprint of RPE1 cells directly after being detached from plate using trypsin (cyan), and after 30 minutes in suspension (magenta). Contours show 95 % density iso-lines, dashed contours 50 % iso-lines. (b) Comparison of deformation medians of N=10 replicates. In interphase the cells show a significant change in deformability due to being kept in suspension for 30 min. The mitotic subpopulation on the other hand, does not change significantly based on t-Test (two-sided, Welch corrected) with **: p<0.01 and n.s.: p≥0.05. Box plot whiskers range from 5th to 95th percentiles, box from 25th to 75th percentile, horizontal line denotes the median, and mean is symbolized by a small square. N=10 replicates with samples from the same cell culture but treated and measured independently on different days, were analyzed.
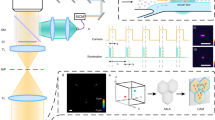
Supplementary Figure 14 Technical details of the alignment light path.
For (a) excitation and (b) emission light paths of the RT-FDC setup. For an explanation of the various parts, see Online Methods, section Experimental Setup.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Discussion and Supplementary Tables 1–5
Source data
Rights and permissions
About this article
Cite this article
Rosendahl, P., Plak, K., Jacobi, A. et al. Real-time fluorescence and deformability cytometry. Nat Methods 15, 355–358 (2018). https://doi.org/10.1038/nmeth.4639
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4639
This article is cited by
-
Experimental measurement and numerical modeling of deformation behavior of breast cancer cells passing through constricted microfluidic channels
Microsystems & Nanoengineering (2024)
-
Fluorescence imaging deformability cytometry: integrating nuclear structure with mechanical phenotyping
Med-X (2023)
-
Rapid single-cell physical phenotyping of mechanically dissociated tissue biopsies
Nature Biomedical Engineering (2023)
-
Machine learning assisted real-time deformability cytometry of CD34+ cells allows to identify patients with myelodysplastic syndromes
Scientific Reports (2022)
-
The impact of cell culture media on the interaction of biopolymer-functionalized gold nanoparticles with cells: mechanical and toxicological properties
Scientific Reports (2022)