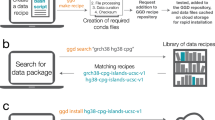
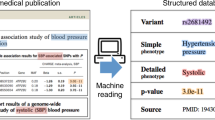
Abstract
GIGGLE is a genomics search engine that identifies and ranks the significance of genomic loci shared between query features and thousands of genome interval files. GIGGLE (https://github.com/ryanlayer/giggle) scales to billions of intervals and is over three orders of magnitude faster than existing methods. Its speed extends the accessibility and utility of resources such as ENCODE, Roadmap Epigenomics, and GTEx by facilitating data integration and hypothesis generation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Quinlan, A.R. & Hall, I.M. Bioinformatics 26, 841–842 (2010).
Li, H. Bioinformatics 27, 718–719 (2011).
Sheffield, N.C. & Bock, C. Bioinformatics 32, 587–589 (2016).
Favorov, A. et al. PLOS Comput. Biol. 8, e1002529 (2012).
Elmasri, R., Wuu, G.T.J. & Kim, Y.-J. The time index: an access structure for temporal data. in Proceedings of the 16th International Conference on Very Large Data Bases (VLDB '90) (eds. McLeod, D., Sacks-Davis, R. & Schek, H.-J.) 1–12 (Morgan Kaufmann, San Francisco, California, USA 1990).
Ernst, J. & Kellis, M. Nat. Methods 9, 215–216 (2012).
Layer, R.M., Skadron, K., Robins, G., Hall, I.M. & Quinlan, A.R. Bioinformatics 29, 1–7 (2013).
De, S., Pedersen, B.S. & Kechris, K. Brief. Bioinform. 15, 919–928 (2014).
Xiao, Y. et al. Bioinformatics 30, 801–807 (2014).
MacQuarrie, K.L. et al. Mol. Cell. Biol. 33, 773–784 (2013).
Farh, K.K.-H. et al. Nature 518, 337–343 (2015).
Mei, S. et al. Nucleic Acids Res. 45, D658–D662 (2017).
Splinter, E. et al. Genes Dev. 20, 2349–2354 (2006).
Nativio, R. et al. PLoS Genet. 5, e1000739 (2009).
Xu, Y. et al. PLoS Genet. 12, e1005992 (2016).
Carroll, J.S. et al. Cell 122, 33–43 (2005).
Theodorou, V., Stark, R., Menon, S. & Carroll, J.S. Genome Res. 23, 12–22 (2013).
Mohammed, H. et al. Nature 523, 313–317 (2015).
Hanstein, B. et al. Proc. Natl. Acad. Sci. USA 93, 11540–11545 (1996).
Li, W. et al. Mol. Cell 59, 188–202 (2015).
Periyasamy, M. et al. Cell Rep. 13, 108–121 (2015).
Mohammed, H. et al. Cell Rep. 3, 342–349 (2013).
Lizio, M. et al. Genome Biol. 16, 22 (2015).
Acknowledgements
We are grateful to the anonymous reviewers for their suggestions and comments. This research was funded by US National Institutes of Health awards to R.M.L. (K99HG009532) and A.R.Q. (R01HG006693, R01GM124355, U24CA209999).
Author information
Authors and Affiliations
Contributions
R.M.L. conceived and designed the study, developed GIGGLE, and wrote the manuscript. B.S.P. developed the GIGGLE score and the PYTHON and GO APIs. T.D. developed the web interface. G.T.M. provided input in the development of the web interface. J.G. conceived and designed the ChIP-seq experiment. A.R.Q. conceived and designed the study and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 GIGGLE indexing process.
(a) Three example annotation sets shown graphically (left) and encoded in files (right) by start position, end position, and ID. (b) GIGGLE's bulk indexing process. (c) The GIGGLE interval search process.
Supplementary Figure 2 The GIGGLE scores for all pairwise combinations of the ChIP-seq datasets for the MCF-7 cell line.
Group 1 highlights the relationship between CTCF, RAD21, and STAG1. Group 2 highlights ERS1, FOXA1, GATA3, and EPS300. Group 3 shows an unexpected relationship between H2AFX and GREB1.
Supplementary Figure 3 A web interface that integrates data from of Roadmap and the UCSC genome browser.
(a) Users specify either a single interval or file to upload as the query, and the server responds with the GIGGLE results from an index in a heatmap. In this case the index is of CHROMHMM prediction from Roadmap. The color of each cell indicates the GIGGLE score, and users can click on a cell (e.g., Myoblast enhancers, marked in red) for more information. (b) When a cell is selected by the user, a window opens that contains the list of intervals in that particular Roadmap cell type/genome state annotation that overlap the query. Each interval is a link that can be followed (e.g., chr1:33642000-33642800, marked in red) for more information. (c) When an interval is selected, that interval becomes a query to a GIGGLE index of the UCSC genome browser tracks. The result gives the set of tracks that contain an interval that overlaps the query, and the web interface opens a window with a “smartview” where only those tracks with overlaps are displayed.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Tables 1–4 (PDF 2119 kb)
Supplementary Data 1
Data used to generate Figure 1. (ZIP 133 kb)
Supplementary Data 2
Data used to generate Figure 2. (ZIP 34860 kb)
Supplementary Data 3
Cell line, tissue, and trait names from Figure 1; accession numbers from Figures 1 and 2 and Supplementary Figure 2. (XLSX 41 kb)
Supplementary Software
GIGGLE source code and experiment scripts. (ZIP 3041 kb)
Rights and permissions
About this article
Cite this article
Layer, R., Pedersen, B., DiSera, T. et al. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods 15, 123–126 (2018). https://doi.org/10.1038/nmeth.4556
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4556
This article is cited by
-
MYC activity at enhancers drives prognostic transcriptional programs through an epigenetic switch
Nature Genetics (2024)
-
An intronic LINE-1 regulates IFNAR1 expression in human immune cells
Mobile DNA (2023)
-
SARS-CoV-2 infection induces epigenetic changes in the LTR69 subfamily of endogenous retroviruses
Mobile DNA (2023)
-
An autoimmune pleiotropic SNP modulates IRF5 alternative promoter usage through ZBTB3-mediated chromatin looping
Nature Communications (2023)
-
FT-6876, a Potent and Selective Inhibitor of CBP/p300, is Active in Preclinical Models of Androgen Receptor-Positive Breast Cancer
Targeted Oncology (2023)