Abstract
Mechanical forces are integral to many biological processes; however, current techniques cannot map the magnitude and direction of piconewton molecular forces. Here, we describe molecular force microscopy, leveraging molecular tension probes and fluorescence polarization microscopy to measure the magnitude and 3D orientation of cellular forces. We mapped the orientation of integrin-based traction forces in mouse fibroblasts and human platelets, revealing alignment between the organization of force-bearing structures and their force orientations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Dembo, M., Oliver, T., Ishihara, A. & Jacobson, K. Biophys. J. 70, 2008–2022 (1996).
Polacheck, W.J. & Chen, C.S. Nat. Methods 13, 415–423 (2016).
Stabley, D.R., Jurchenko, C., Marshall, S.S. & Salaita, K.S. Nat. Methods 9, 64–67 (2011).
Liu, Y., Galior, k., Ma, Y.P.-Y. & Salaita, K. Accounts Chem. Res. https://doi.org/10.1021/acs.accounts.7b00305 (2017).
Zhang, Y., Ge, C., Zhu, C. & Salaita, K. Nat. Commun. 5, 5167 (2014).
Liu, Y. et al. Proc. Natl. Acad. Sci. USA 113, 5610–5615 (2016).
Grashoff, C. et al. Nature 466, 263–266 (2010).
Iqbal, A. et al. Proc. Natl. Acad. Sci. USA 105, 11176–11181 (2008).
Qiu, Y. et al. Proc. Natl. Acad. Sci. USA 111, 14430–14435 (2014).
Schwarz Henriques, S., Sandmann, R., Strate, A. & Köster, S. J. Cell Sci. 125, 3914–3920 (2012).
Coller, B.S. & Shattil, S.J. Blood 112, 3011–3025 (2008).
Kampmann, M., Atkinson, C.E., Mattheyses, A.L. & Simon, S.M. Nat. Struct. Mol. Biol. 18, 643–649 (2011).
DeMay, B.S., Noda, N., Gladfelter, A.S. & Oldenbourg, R. Biophys. J. 101, 985–994 (2011).
Diagouraga, B. et al. J. Cell Biol. 204, 177–185 (2014).
Sunyer, R. et al. Science 353, 1157–1161 (2016).
Tang, X., Tofangchi, A., Anand, S.V. & Saif, T.A. PLoS Comput. Biol. 10, e1003631 (2014).
Weisel, J.W. Biophys. Chem. 112, 267–276 (2004).
Zhang, Y. et al. Proc. Natl. Acad. Sci. USA. https://doi.org/10.10731/pnas.1710828115 (2017).
Legant, W.R. et al. Proc. Natl. Acad. Sci. USA 110, 881–886 (2013).
Kanchanawong, P. et al. Nature 468, 580–584 (2010).
Paszek, M.J. et al. Nat. Methods 9, 825–827 (2012).
Swaminathan, V. et al. Proc. Natl. Acad. Sci. USA 114, 10648–10653 (2017).
Zhu, J. et al. Mol. Cell 32, 849–861 (2008).
Axelrod, D. Biophys. J. 26, 557–573 (1979).
Chan, T.F. & Vese, L.A. IEEE Trans. Image Process. 10, 266–277 (2001).
Galush, W.J., Nye, J.A. & Groves, J.T. Biophys. J. 95, 2512–2519 (2008).
Vacklin, H.P., Tiberg, F. & Thomas, R.K. Biochim. Biophys. Acta 1668, 17–24 (2005).
Neese, F. Comp. Mol. Sci. 2, 73–78 (2012).
Becke, A.D. J. Chem. Phys. 98, 1372–1377 (1993).
Lee, C., Yang, W. & Parr, R.G. Phys. Rev. B Condens. Matter 37, 785–789 (1988).
Vosko, S.H., Wilk, L. & Nusair, M. Can. J. Phys. 58, 1200–1211 (1980).
Stephens, P.J., Devlin, F.J., Chablowski, C.F. & Frisch, M.J. J. Phys. Chem. 98, 11623–11627 (1994).
Krishnan, R., Binkley, J.S., Seeger, R. & Pople, J.A. J. Chem. Phys. 72, 650–654 (1980).
Chai, J.-D. & Head-Gordon, M. J. Chem. Phys. 128, 084106 (2008).
Dunning, T.H. Jr. J. Chem. Phys. 90, 1007–1023 (1989).
Klamt, A. J. Phys. Chem. 99, 2224–2235 (1995).
Marenich, A.V., Cramer, C.J. & Truhlar, D.G. J. Phys. Chem. B 113, 6378–6396 (2009).
Acknowledgements
The authors thank A. Garcia (Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University) for providing NIH-3T3 fibroblasts stably transfected with GFP-paxillin; E. Bartle and T. Urner for assistance with optical systems; and Y. Liu and J. Kindt for helpful discussion. The work was supported through NIGMS R01 GM124472 (K.S.), NSF 1350829 (K.S.), NSF CAREER 1553344 (A.L.M.), NSF IDBR 1353939 (K.S. and A.L.M.), NSF GRFP DGE-1444932 (J.M.B., A.T.B., W.D.D., and M.E.F.), NCI 1F99CA223074-01 (V.P.-Y.M.), Alfred P. Sloan Foundation (F.A.E.), NSF CAREER 1150235 (W.A.L.), NIH Grants RO1HL121264 (W.A.L.), RO1 HL130918 (W.A.L.), U01-HL117721 (W.A.L.), and U54HL112309 (W.A.L.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Contributions
A.L.M. and K.S. conceived the general principle of MFM. J.M.B., A.T.B., A.L.M., and K.S. conceived of applying excitation-resolved fluorescence microscopy. J.M.B., A.T.B., A.L.M., and K.S. designed experiments. J.M.B. performed experiments. J.M.B. performed anisotropy simulations. A.T.B. derived tilt-angle measurement and performed error simulations. J.M.B., Y.Z., and V.P.-Y.M. synthesized reagents and prepared DNA-modified surfaces. J.M.B. and A.T.B. performed data analysis and wrote the required programs. J.M.B., M.E.F., and W.A.L. designed platelet aggregate experiments. M.E.F. assisted with platelet handling. W.D.D. and F.A.E. calculated the Cy3B transition dipole moment. J.M.B., A.T.B., A.L.M., and K.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 HPLC and MALDI-TOF Characterization of MFM Probes
(a) Synthesis scheme for MFM probes. HPLC chromatogram and MALDI-TOF spectra for (b) Product 1, (c) Product 2, (d) Product 3, (e) Product 4, (f) Product 5, and (g) Product 6.
Supplementary Figure 2 Receptor Force Confines the Mobility of DNA Tension Probes.
(a) The DNA tension probe is composed of 2 dsDNA segments, (length 7 nm << lp, dsDNA = 53 nm) which we treat as rigid rods. Flexible ssDNA links these rods. Rotatable bonds are indicated by red dashed circles. (b) The dsDNA rod may rotate angle β from its equilibrium position (arc of motion is indicated by the green arc). The cyanine fluorophore (yellow disc) can theoretically adopt any plane tangential to the arc. The motion of the dsDNA rod along this arc is constrained by its attachment to the ssDNA entropic spring. We assume that the integrin and the bottom of the ssDNA are fixed in place, therefore any extension of the ssDNA produces a rotation about the integrin. Receptor force is sufficient to produce an extension, x0, of the ssDNA spring (black curvy line), but extending the hairpin further, to an extension x’ (red curvy line) requires additional energy input. This energy must come from the thermal energy of the system. (c) To illustrate the full geometry, we have superimposed the constrained arc of motion of the dsDNA upper arm of the construct onto the scheme from Figure 1b in the main text. For clarity, the entire DNA tension probe (two dsDNA rods and the ssDNA hairpin) is represented as a single blue cylinder as in the main text. The green arc illustrates the flexibility of the full DNA tension probe. (d) Average angle between the dsDNA helix and the axis of cellular force (β) as a function of applied cellular force. (e) Error in the measurement of θforce and (f) Φforce due to the flexibility of the ssDNA portion of the DNA tension probe. For e and f, β angles were randomly chosen by binning 100 random numbers on the interval 0-1 into the CDF for β. Each pixel in e and f represents the average error of 100 β angles. This stochastic simulation was repeated 3 times with similar results.
Supplementary Figure 3 Light Paths
(a) Emission resolved and (b) Excitation resolved fluorescence polarization microscopy optical paths. For both set-ups, the laser was operated in widefield mode.
Supplementary Figure 4 Cy3B-DNA Stacking Enables Fluorescence Anisotropy Measurement of Platelet Traction Forces
(a) Cy3B linked to the MFM probe via a 6-carbon linker (0T) exhibited increased fluorescence anisotropy (median 0.257) compared to soluble Cy3B (median 0.057). Adding 2 additional thymines (2T) to the linker reduced the anisotropy (median 0.191), despite the increased mass. Alexa 488, a hydrophilic dye not expected to stack with DNA, had low fluorescence anisotropy when linked to the MFM probe (median 0.066). All differences are statistically significant (2 tailed 2 sample t-test on the sample means of n=3 independent experiments, 6 measurements per experiment: 0T Cy3B-2T Cy3B p=5.2x10−4, 95% confidence interval 0.052-0.089; 0T Cy3B-Cy3B p=8.6x10−7, 95% confidence interval 0.19-0.21; 2T Cy3B-Cy3B p=4.5x10−5, 95% confidence interval 0.11-0.15). The red line indicates the median, black dots represent the sample means, 25/75% quartiles are indicated by the blue box, whiskers extend to 1.5 times the interquartile range). (b) Platelets are expected to be roughly contractile, orienting MFM probes centripetally. (c) Geometry of the anisotropy model described in Supplementary Note 2. (d) The predicted pattern of anisotropy fits a cos2α function. (e) Measurements of platelets and the 0T Cy3B probe revealed an anisotropy pattern resembling a bowtie. Splitting the anisotropy into wedges (0° indicated in panel b) revealed the expected cos2α distribution. We measured the amplitude of the fitted cos2 angular anisotropy distributions for many platelets. The amplitude was attenuated for 2T and Alexa 488 tension probes compared to the 0T probe (0T, median 0.086, 107 platelets, n=4 independent experiments; 2T, median 0.049, 116 platelets, n=4 experiments; Alexa 488, median 0.028, 71 platelets, n=3 independent experiments). All differences are statistically significant (2 tailed 2 sample t-test allowing unequal variance on the amplitudes of individual platelets: 0T Cy3B-2T Cy3B, p=3.3x10−18, 95% confidence interval 0.026-0.040; 0T Cy3B-Alexa 488, p=4.1x10−30, 95% confidence interval 0.045-0.060; 2T Cy3B-Alexa 488, p=5x10−9, 95% confidence interval 0.013-0.025). The red line indicates the median, 25/75% quartiles are indicated by the blue box, whiskers extend to 1.5 times the interquartile range, and outliers are indicated by red dots). (f) Representative images of RICM, tension, and anisotropy for a platelet on a 0T Cy3B DNA probe modified surface. The white arrow indicates the excitation polarization. (g) The anisotropy pattern for the platelet from Supplementary Fig. 4f when the excitation polarization is rotated 90°. (f) Representative 2T Cy3B platelet anisotropy. (g) Representative Alexa 488 platelet anisotropy. The average anisotropy plots show the mean anisotropy value within 16 evenly spaced wedges rotating α degrees around the center of each of the 4 indicated platelets. Please note that the platelet shown in Supplementary Fig. 4f and 4g is also featured in Figure 1c). All experiments were performed with the 4.7 pN hairpin.
Supplementary Figure 5 Computing θforce from Excitation Resolved Fluorescence Polarization Experiments.
(a) Coordinate system and (b) diagram of fluorescence excitation event. All vectors are given a magnitude of 1. The orange vector represents the transition dipole moment of the fluorophore. The green vector represents the electric field vector E of plane-polarized light used to excite the fluorophore. The blue vector represents the DNA helix vector h, which is equivalent to the force vector (shown in panel a) and is specified by θforce and Φforce. (c) We generated a discrete approximation set of permissible fluorophore orientations when μ is stacked orthogonally to h. (d) We sought to validate that Cy3B’s TDM lies in the plane of the dye. We computed the molecular orbitals involved in the first absorption and emission excitation in Cy3B calculated using Time-Dependent Density Functional Theory (TDDFT) at the B3LYP/6-31G* level of theory. Orbitals are plotted using Visual Molecular Dynamics software package (VMD, version 1.9.1) with an isosurface contour value of 0.05. The ground state geometry of Cy3B optimized in water using the conductor-like-screening solvation model (COSMO) model in the ORCA software package (left) and the optimized excited state geometry of Cy3B (right) are both shown. (e) Optimized ground state geometry (left, S0; Absorption Dipole) and excited state geometry (right, S1; Emission Dipole, shown on the right) overlaid with the calculated absorption and emission dipoles (red arrows). The red arrows represent the TDM for absorption (left) and emission (right) respectively. Note that the absorption and emission dipoles are essentially parallel and that both lie in the plane of the cyanine fluorophore. (f) We generated the minimum and maximum Pexc value as a function of θforce while holding Φforce and Φexcitation constant. The minimum excitation probability (Pexc,min) varies as a function of θforce. (g) The maximum excitation probability (Pexc,max) does not vary as a function of θforce. (h) The ratio Q (Pexc,min/ Pexc,max) is readily measured experimentally and varies as a function of tilt angle. (i) The assumption that fluorophores are isotropic emitters introduces no more than 2.5° error in the measured tilt angle (θforce,measured, elsewhere denoted as θforce).
Supplementary Figure 6 MFM Accuracy Depends on Signal-Noise Ratio and Ensemble Averaging
Error due to noise was estimated using a Monte Carlo method. Expected intensity values for 72 evenly spaced Φexcitation values between 0 and 180° for a given helix orientation were generated using methods described in Supplementary Note 3 (in this case, I=Pexc*Imax). Normally distributed background noise σbkrd (denoted backgroundsurface in equation 3), as well as photon noise σpho=√I was then added to each measurement. θforce and Φforce were estimated using the noise-added intensity values and the method described in this paper. The errors in θforce and Φforce, denoted ɛθ and ɛΦ, respectively, were calculated as the absolute difference between the true values and the calculated values of θforce and Φforce. When calculating photon noise, we assumed that one photon produced one count on the detector. This process was repeated 100 times for a given orientation, and the average error was calculated. ɛθ and ɛΦ are displayed as heat maps. (a) Error depends on θforce, but not on Φforce for all three levels of background noise tested. (b) ɛΦ is high for small tilt angles and poor signal to noise ratios. θforce is overestimated for poor signal to noise ratios and low values of θforce. (c) Cross sections across the heatmaps in b at a simulated intensity of 1000 a.u. enable easy visualization of the dependence of ɛθ and ɛΦ on θforce and signal to noise ratio. (d) Schematic representation of two hairpins that differ in the Z component of the cellular force (indicated by the green double-headed arrow). As in Figure 1b, fluorophore orientations are rendered as colored discs while the XY projection of each disk is rendered as a translucent ellipse in the XY plane. The red and yellow hairpins produce the red and yellow fluorescence intensity curves as a function of Φexcitation. Adding the fluorescence intensity curves and renormalizing to the new maximum intensity reveals the resultant sinusoid (blue boxes). For the case where the red and yellow hairpins differ in the force Z-component, the measurement of Φforce is not altered; however, ensemble averaging does produce an intermediate amplitude sinusoid (blue boxes). (e) Schematic representation of two hairpins that differ in the XY orientation of applied receptor force (indicated by the blue double-headed arrow). Adding these curves produces a new sinusoid with phase (Φforce) that is the average of the phases of the component sinusoids. The amplitude of the resultant sinusoid is also reduced, causing underestimation of the tilt angle (θforce).
Supplementary Figure 7 Analysis of Background Signal
(a) A representative tension signal image of platelets engaged with a tension sensor surface. We defined two ROI’s including a platelet (ROI 1) and the background (ROI 2). (b) Histogram plot displaying the raw fluorescence intensity of the ROI 2 (background, orange) and the platelet signal (tension, blue). Note that the tension signal (from ROI 1) is masked by only including fluorescence with a signal to noise ratio of 5 and greater. (c) A scatter plot of tilt angle versus intensity for platelet tension and the background. The background tilt angle is disordered, spanning the range 0-45° with an average of 18° while the tilt angle for this platelet spans the range 0-67° with a mean of 38°. (d) Scatter plot of Φforce for the background shows a broad range of angles that span -90° to 90°. (e) Representative raw data for platelet tension and background along with their respective fits (amplitude 160 A.U. for platelet, 7.8 A.U. for background). Note that the amplitude of the fit is an important parameter for MFM, as it defines the tilt angle and our ability to measure Φforce accurately. The amplitude for the platelet signal is 20 fold greater than the background. (f) Magnified view of the background raw data and fit shown in (e) to display the error-prone nature of curve fitting the background signal. The data shown is representative of background analysis of 3 images from each of n=3 independent platelet and fibroblast experiments.
Supplementary Figure 8 Fluorescence Polarization Measurements of DiI Coated Beads
(a) 5 μm silica beads were coated with 100% DOPC supported lipid bilayers (SLB) doped with DiI, a dye that inserts carbon tails into lipid membranes. The TDM of DiI is therefore parallel to the SLB/bead surface [Badley, R.A., Martin, W.G. & Schneider, H. Biochemistry 12, 268-275 (1973)]. (b) Fluorescence anisotropy measurements reveal a cos2 distribution consistent with TDMs aligned parallel to the bead surface. The anisotropy pattern is representative of 42 beads (from n=3 independent experiments). (c) Rotating the excitation polarization (green dashed line) results in observable fluorescence intensity changes. The DiI fluorescence is most intense on the edges of the bead that are parallel to the excitation polarization. (d) Plotting the fluorescence intensity of a pixel as a function of excitation polarization reveals a cos2θ distribution. The excitation polarization angle of the peak fluorescence, the azimuth, corresponds to the fluorophore TDM orientation in that pixel. (e) DiI azimuths are tangential to the surface of the bead as expected, demonstrating the accuracy of our optical system in measuring the orientation of fluorophores via excitation-resolved fluorescence polarization. The data shown is representative of 70 beads (n=3 independent experiments, 2 experiments were performed with the 73-image acquisition, one with a previously reported 4-point acquisition [DeMay, B.S., Noda, N., Gladfelter, A.S. & Oldenbourg, R. Biophys J 101, 985-994 (2011)]).
Supplementary Figure 9 Multiple MFM Maps
(a) Platelet MFM maps with radial histograms and average force axes displayed as black bars on the radial histograms. Grey background represents pixels below an intensity threshold (signal-noise ratio < 5). These images are representative of 79 total platelet MFM maps from n=5 independent experiments. (b) Platelet clot MFM map with platelet borders indicated by the white borders and average platelet force orientations denoted by the double headed white arrows. This image is representative of 6 platelet aggregate MFM maps from n=3 independent experiments. (c) Fibroblast MFM map with radial histograms and average force axis displayed as a black bar on the radial histograms. These measurements are representative of 37 fibroblast MFM maps from n=3 independent experiments. All measurements were performed with the 4.7 pN hairpin. Grey background represents pixels below an intensity threshold (signal-noise ratio < 5).
Supplementary Figure 10 Platelet x-y Force Distributions Suggest Two Patterns of Platelet Traction Forces
Sample radial histograms of platelet XY force axes (a random subset of the 79 analyzed platelets from n=5 independent experiments) reveal two populations. We quantify these populations by calculating the circular variance of platelet XY forces. (a) In low circular variance platelets (variance < 0.65 based on the bimodal distribution from Fig. 1i), XY forces are concentrated in a few bins within the rose plots. (b) In high circular variance platelets (variance > 0.65), XY forces appear more uniformly distributed.
Supplementary Figure 11 Platelet Immunostaining
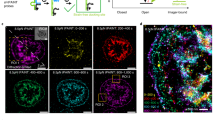
Platelets were fixed and stained for actin and tubulin. Tension (measured before fixation) colocalized with actin and anti-localized with tubulin. Platelet tension was imaged in conjunction with actin in n=2 independent experiments. Platelet actin and tubulin were co-stained in n=3 independent experiments. Platelet tension, actin, and tubulin (shown here) were simultaneously imaged in 52 platelets (from one independent experiment).
Supplementary Figure 12 Two Spatially Distinct Regions of Platelet Force Orientation
(a) RICM and (b) 4.7 pN tension signal for a representative human platelet. (c) The angle between the measured force orientation and a vector oriented towards the cell’s centroid was computed and displayed as a heatmap. To quantify the behavior of the contractile ring compared to the interior tension signal, masks were generated from the RICM image via a Chan-Vese edge finding algorithm. (d) Masks were generated of the outermost 7 pixels, the “Ring” and (e) the “Center” regions of each platelet. (f) The deviation from isotropic force alignment within the center and ring of the platelet shown above were quantified. The ring median deviation is 10.7° deviation with 1057 pixels analyzed. Center median deviation is 16.25° with 3035 pixels analyzed. The red line indicates the median, quartiles are indicated by the blue box, whiskers extend to 1.5 times the interquartile range, and the red dots indicate outliers. Note that adjacent pixels are not independent measurements, thus statistical tests were not employed here. (g) The average deviation from isotropic force alignment was quantified for all platelets, revealing that platelet traction force alignment is different in the center compared to the exterior ring (the average center and ring deviation from isotropic force alignment of 79 platelets from n=5 experiments were pooled for a paired t-test, p=2.2x10−8, 95% confidence interval -6.6° to -3.4°). Ring median deviation is 16.0°. Center median deviation is 21.3°. The red line indicates the median, quartiles are indicated by the blue box, whiskers extend to 1.5 times the interquartile range, and the red dots indicate outliers.
Supplementary Figure 13 Focal Adhesion Force Analysis
(a) We identified focal adhesions (FA) by segmenting the 3T3 cell tension images. 494 focal adhesions were identified from 15 single fibroblasts entirely contained within the image frame (n=3 experiments). (b) For each FA, we measured the average force axis and calculated an axis oriented from the FA centroid to the centroid of the cell (the centripetal axis). (c) For individual FAs, the deviation between the average FA force axis and the centripetal axis was only 21.3° and the distribution was heavily biased towards 0° deviation, indicating general force alignment along axes oriented towards the cell center. (d) The circular variance for individual FA’s was low (average circular variance of 0.236), indicating coherent force alignment. (e) We measured the difference between the distal and proximal halves of FAs and found this difference was small (1.5°), suggesting that integrin forces FA forces normal to the substrate are also tightly organized.
Supplementary Figure 14 Comparison of Platelet and Fibroblast Tilt Angles
(a) Histograms of pixel-by-pixel tilt angles observed in platelets and fibroblasts reveal similar whole cell mean tilt angles: platelets, 40°±2°; fibroblasts, 39°±4° (mean ± standard deviation). The means are not significantly different (2 tailed 2 sample t-test allowing unequal variance, p=0.09, 95% confidence interval -0.0034-0.043; platelet data pooled from 79 platelets from n=5 experiments; fibroblast data pooled from 37 cells from n=3 experiments). (b) Platelet tilt angle varies little from the cell center (median 34.7°) to the cell edge (median 39.9°) while fibroblast tilt angles increase from the cell center (median 28.7°) compared to at the cell edge (median 41.3°). The data were pooled from 79 platelets from n=5 experiments and 15 well-spread, single fibroblasts entirely contained within the image frame from n=3 experiments. The red lines indicate medians. The 25/75% quartiles are indicated by blue boxes, while whiskers extend to 1.5 times the interquartile range. Outliers are indicated by red “+” signs. The box plot displays the average tilt angle of all pixels within the specified radius from the cell center. 79 platelets were included in every bin but because not all fibroblasts had FAs near the cell center, 5 fibroblasts were included in the 0.1 bin, 11 fibroblasts were included in the 0.3 bin, and 15 fibroblasts were included in bins 0.5-0.9. A one-way ANOVA was used to test the significance in the variation of θforce as a function of normalized radius. Both platelet and fibroblast θforce variation are significant (platelet, p=5x10−19, F=25.86; fibroblast, p=3.2x10−7, F=12.25) (c) Scheme depicting the tilt angle trends for platelets and fibroblasts.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Notes 1–4 and Supplementary Tables 1–4 (PDF 3454 kb)
Life Sciences Reporting Summary
Life Sciences Reporting Summary (PDF 159 kb)
Supplementary Software 1
MFM GUI (ZIP 4890 kb)
Supplementary Software 2
Analysis Script (ZIP 14 kb)
Rights and permissions
About this article
Cite this article
Brockman, J., Blanchard, A., Pui-Yan, V. et al. Mapping the 3D orientation of piconewton integrin traction forces. Nat Methods 15, 115–118 (2018). https://doi.org/10.1038/nmeth.4536
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4536
This article is cited by
-
DNA mechanocapsules for programmable piconewton responsive drug delivery
Nature Communications (2024)
-
Can a bulky glycocalyx promote catch bonding in early integrin adhesion? Perhaps a bit
Biomechanics and Modeling in Mechanobiology (2024)
-
Molecular mechanocytometry using tension-activated cell tagging
Nature Methods (2023)
-
Organization, dynamics and mechanoregulation of integrin-mediated cell–ECM adhesions
Nature Reviews Molecular Cell Biology (2023)
-
Detection of cellular traction forces via the force-triggered Cas12a-mediated catalytic cleavage of a fluorogenic reporter strand
Nature Biomedical Engineering (2023)