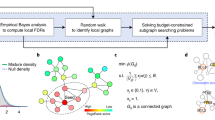
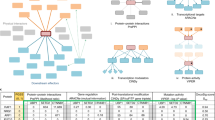
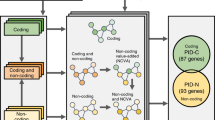
Abstract
Methods that integrate molecular network information and tumor genome data could complement gene-based statistical tests to identify likely new cancer genes; but such approaches are challenging to validate at scale, and their predictive value remains unclear. We developed a robust statistic (NetSig) that integrates protein interaction networks with data from 4,742 tumor exomes. NetSig can accurately classify known driver genes in 60% of tested tumor types and predicts 62 new driver candidates. Using a quantitative experimental framework to determine in vivo tumorigenic potential in mice, we found that NetSig candidates induce tumors at rates that are comparable to those of known oncogenes and are ten-fold higher than those of random genes. By reanalyzing nine tumor-inducing NetSig candidates in 242 patients with oncogene-negative lung adenocarcinomas, we find that two (AKT2 and TFDP2) are significantly amplified. Our study presents a scalable integrated computational and experimental workflow to expand discovery from cancer genomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
19 December 2017
In the version of this article initially published online, the color labels for oncogene-positive and oncogene-negative lung adenocarcinomas were swapped in the Figure 3a legend. The error has been corrected in the print, PDF and HTML versions of this article.
References
Garraway, L.A. & Lander, E.S. Lessons from the cancer genome. Cell 153, 17–37 (2013).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Frampton, G.M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Roychowdhury, S. et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci. Transl. Med. 3, 111ra121 (2011).
Van Allen, E.M. et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 4, 1140–1153 (2014).
Wagle, N. et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2, 82–93 (2012).
Gonzalez-Perez, A. & Lopez-Bigas, N. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 40, e169 (2012).
Mermel, C.H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Taylor, B.S. et al. Functional copy-number alterations in cancer. PLoS One 3, e3179 (2008).
Lohr, J.G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 109, 3879–3884 (2012).
Lawrence, M.S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Ciriello, G., Cerami, E., Sander, C. & Schultz, N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 22, 398–406 (2012).
Hofree, M., Shen, J.P., Carter, H., Gross, A. & Ideker, T. Network-based stratification of tumor mutations. Nat. Methods 10, 1108–1115 (2013).
Leiserson, M.D.M. et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 47, 106–114 (2015).
Vandin, F., Upfal, E. & Raphael, B.J. Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol. 18, 507–522 (2011).
Babur, Ö. et al. Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations. Genome Biol. 16, 45 (2015).
Miller, C.A., Settle, S.H., Sulman, E.P., Aldape, K.D. & Milosavljevic, A. Discovering functional modules by identifying recurrent and mutually exclusive mutational patterns in tumors. BMC Med. Genomics 4, 34 (2011).
Yeang, C.-H., McCormick, F. & Levine, A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J. 22, 2605–2622 (2008).
Creixell, P. et al. Pathway and network analysis of cancer genomes. Nat. Methods 12, 615–621 (2015).
Lage, K. et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25, 309–316 (2007).
Li, T. et al. A scored human protein–protein interaction network to catalyze genomic interpretation. Nat. Methods 14, 61–64 (2016).
Berger, A.H. et al. High-throughput phenotyping of lung cancer somatic mutations. Cancer Cell 30, 214–228 (2016).
Boehm, J.S. et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 (2007).
Dunn, G.P. et al. In vivo multiplexed interrogation of amplified genes identifies GAB2 as an ovarian cancer oncogene. Proc. Natl. Acad. Sci. USA 111, 1102–1107 (2014).
Kim, E. et al. Systematic functional interrogation of rare cancer variants identifies oncogenic alleles. Cancer Discov. 6, 714–726 (2016).
Campbell, J.D. et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48, 607–616 (2016).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012).
Marbach, D. et al. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat. Methods 13, 366–370 (2016).
Lage, K. et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl. Acad. Sci. USA 105, 20870–20875 (2008).
Rossin, E.J. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Carter, S.L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Acknowledgements
J.D.C. is supported by the LUNGevity Career Development Award (CDA). H.H. was supported by a Fund for Medical Discovery Award from the Executive Committee On Research at Massachusetts General Hospital. H.H. and K.L. are supported by the MGH IRG American Cancer Society. K.L. is supported by a grant from the Stanley Center at the Broad Institute, a Broadnext10 grant from the Broad Institute, 1R01MH109903, a Large Thematic Project Grant from the Lundbeck Foundation (R223-2016-721), and a Research Award from the Simons Foundation (SFARI).
Author information
Authors and Affiliations
Contributions
H.H. developed, benchmarked, and implemented the NetSig algorithm with input from M.S.L. and supervision from G.G. and K.L. C.R.C., Y.S., E.S., N.I., and E.K. executed the in vivo tumorigenesis experiments with input from H.H. and K.L. and supervision from J.S.B. H.H. developed and implemented the quantitative analytical framework of in vivo tumorigenesis data with input from C.R.C., Y.S., and E.S. as well as supervision from J.S.B. and K.L. J.D.C. reanalyzed lung adenocarcinoma data with input from H.H., J.S.B., G.G., and K.L. All authors analyzed data and discussed the results. H.H., W.C.H., J.D.C., J.S.B., G.G., and K.L. wrote the manuscript with input from all authors. J.S.B., G.G., and K.L. designed and directed the work. K.L. initiated and led the study.
Corresponding authors
Ethics declarations
Competing interests
K.L. is on the scientific advisory board and is the founder of Intomics A/S with equity in the company. InWeb_InBioMap is a product of Intomics A/S that is freely available to academic users from http://lagelab.org/resources and http://www.intomics.com/inbio/map.
Integrated supplementary information
Supplementary Figure 1 Significance of AUCs reported in the main paper and comparisons to alternative NetSig approaches
Definition of gene sets: We curated four tiers of genes linked to cancer and one tier of randomly chosen genes for control purposes (Supplementary Table 1). Tier 1 (termed ‘Cosmic classic’ in the Main Text and Fig. 1) consists of established (or classic) cancer genes from the Catalogue of Somatic Mutations in Cancer (Cosmic, e.g., TP53, BRCA1, and BRAF). Tier 2 contains genes that have been recently identified as cancer genes from the Sanger Gene Census dataset (e.g., MLL2, CDK12, and GATA2). Tier 3 (termed ‘recently emerging cancer genes’ in the Main Text and Fig. 1) are emerging cancer genes, meaning they have been identified using conservative statistics in cancer sequencing studies, but where the biological connection to known cancer pathways is often unclear (e.g., ING1). Tier 4 contains suspected cancer genes with solid, but in some cases not entirely conclusive, statistical evidence from cancer sequencing studies (e.g., EIF2S2). Tier 5 (termed ‘random genes’ in the Main Text and Fig. 1) is a set of random genes included as a random control in our analysis. Grey squares and triangles indicate the AUC of the relevant Tier when the NetSig classifier is calculated using Q values or P values of pan-cancer MutSig significances, respectively. Grey circles indicate the AUC of the relevant Tier when the influence of Tier 1 (Cosmic classic) genes is removed from the analysis by artificially setting Tier 1 (Cosmic classic) genes to Q = 1 and running the NetSig calculation. The boxes shows the distribution of AUCs observed when NetSig scores are calculated in 100 randomized networks using q values of MutSig pan-cancer significances. Boxes indicate median, first and third quartile of the AUC distributions for the relevant tiers.
Supplementary Figure 2 Testing NetSig across 21 tumor types
In this figure, each box illustrates the results from the tumor-specific NetSig analyses of 17 tumor types that have at least four defined driver genes (first 17 boxes) or for pan-cancer genes (last box). In each plot (for example for breast cancer [BRCA, second box]), the AUC calculated with the NetSig score corresponding to mutation burdens from the tumor in question (NetSigBRCA) is indicated by the colored (in the case of BRCA, orange) line. The ability to distinguish BRCA driver genes using a NetSig score derived from pan-cancer data is indicated with a dark grey curve. The difference in performance using NetSigpan-cancer and NetSigBRCA (in this case 0.76-0.77 = - 0.01) is indicated above the plot as the differential AUC (or dAUC). In the case of BRCA the pan-cancer data is slightly better at classifying BRCA driver genes than the BRCA-specific mutation data.
Supplementary Figure 3 Quantile-quantile plot of observed versus expected NetSig P when correcting for ‘knowledge contamination’
Clockwise from the upper left panel: Quantile-quantile (QQ) when using all MutSig Q values for the NetSig calculation. Upper right panel: Removing the effect of ‘Cosmic classic’ genes by setting their Q to 1 in the NetSig calculation. Lower left pane: Removing the effect of ‘Cosmic classic’ and ‘recently emerging’ cancer genes by setting their Q value to 1 in the NetSig calculation. Lower right panel: QQ plot when running NetSig on a randomized network. The random network shows no inflation, showing that the NetSig method itself does not contribute to inflation.
Supplementary Figure 4 There is no correlation between degree, NetSig significance, and tier membership
We plotted the connectivity of each gene (number of interacting proteins) against the resulting NetSig significances (nominal P values). This shows that neither connectivity nor tier membership is correlating with an index gene’s degree in InWeb (i.e., the amount of genes in its first order network). This observation supports that study bias or “knowledge contamination” is not driving our results. The genes in Tiers 1 - 5 are defined in Supplementary Figure 1.
Supplementary Figure 5 Examples of NetSig5000 genes
The first order network of a) AFF2 (dark grey, NetSig Q = 0.07), b) PIK3CB (dark grey, NetSig Q = 0.016), c) E2F4 (dark grey, NetSig Q = 0.03), d) RUNX2 (dark grey, NetSig Q = 0.07), and e) MYO7A (dark grey, NetSig Q = 0.06) that are significant in the NetSig analysis. Large nodes other than AFF2, E2F4, PIK3CB, RUNX2, and MYO7A are colored by the significance of the pan-cancer Q value of the corresponding gene, where light grey or no shading represents q close to 1 and red Q << 1, with darker red representing more significant Q values. Small nodes represent genes with Q = 1 (or genes not annotated) in the pan-cancer data. All networks can be visualized from www.lagelab.org/resources.
Supplementary Figure 6 Quantile-quantile plot of observed versus expected P values for the lung adenoma carcinoma amplification analysis
The quantile-quantile plot was generated based on all genes that were measured and quantified on the SNP6 array.
Supplementary Figure 7 Testing for enrichment of amplification and single nucleotide variants in oncogene negative samples
Testing for the enrichment of a) gene amplification and b) SSNVs/Indels in genes for oncogene negative samples. Oncogene negative patients were defined as those having no known driver mutation in the RAS/RAF/receptor tyrosine kinase (RTK) pathway. We created a null distribution of P values using Fisher’s exact test on 100 random gene sets (while controlling for overall connectivity in protein-protein interaction space), which is shown as bar plots. The observed p value of data from Figure 3, main text is indicated as the red line. The P values from the data in Figure 3 are better in 96 of a 100 random sets for the amplifications (P = 0.04). This is not the case for the SSNVs and indels (P = 0.19).
Supplementary Figure 8 NetSig candidates representing two conceptual groups
Group 1: a) AKT is a kinase in the PIK3 pathway, which induced tumors when overexpressed in the experiments (i.e., TumorPlex assay). It is also significantly amplified in lung adenocarcinoma samples negative for known driver mutations. b) PIK3CB is a known kinase in the PI3K pathway has been recently shown to contain tumorigenic alleles19 c) PIK3CG is a known kinase in the PI3K pathway that induces tumors in the experiments (i.e., TumorPlex assay). d) and e) RASGRP1 and RASGRP3 both inducted tumors in the experiments (i.e., TumorPlex assay) when overexpressed and are known guanine nucleotide exchange factors in the RAS pathway. Group 2: f) TFDP2 was not earlier known to be involved in cancer and the mechanism by which it is tumorigenic remains unclear. It induces tumors when overexpressed in the experiments (i.e., TumorPlex assay) and is significantly amplified in driver-gene-negative samples in lung adenocarcinoma g) MYO7A has significantly more damaging than benign mutations in patients without known driver mutations from the paper by Lawrence et al.1.
Supplementary Figure 9 Cancer patients without established driver mutations are enriched for deleterious mutations in NetSig5000 genes
a) We compared the fraction of genes with damaging (i.e, probably damaging and possibly damaging pooled into one set) versus benign mutations as determined by PolyPhen in the NetSig5000 genes (on the background of all genes in the genome), and show a statistically significant enrichment of damaging mutations in the NetSig5000 set (P = 0.016, using Fischer’s exact test, NetSig5000 is indicated by dark red and all genes in the genome by light red). b) Using PolyPhen2, we transformed all mutations observed in the NetSig5000 set to continuous normalized scores of how much the mutation is predicted to affect gene function negatively (less damaging to more damaging oriented left to right on the x-axis). When comparing to all genes in the genome, mutations in the NetSig5000 genes are significantly depleted for less damaging PolyPhen scores, and significantly enriched for more damaging PolyPhen scores (P = 0.046, using a non-parametric two-sample Kolmogorov-Smirnov test, histograms show the binned proportions, the line the cumulative distributions of scores). For comparison, we show the results for the same tests run on the Cancer5000 set in panels c) and d), respectively (with Cancer5000 significant genes in dark blue and the background genes in light blue). While the trends and proportions of deleterious versus benign mutations observed in the Cancer5000 genes are similar to our observations for the NetSig5000 genes (thus supporting the cancer relevance of the NetSig5000 set), the statistical significances levels are higher due to more genes in the Cancer5000 set and because the effect size, as expected, is larger.
Supplementary Figure 10 Distribution of damaging to benign mutation ratios from patients with no established driver mutation in candidate genes from Supplementary Figure 7 compared to a random expectation
For genes from Supplementary Figure 7 we calculated the ratio of damaging to benign mutations (indicated by a diamond, raw data in Supplementary Table 8 and 9). We compared this ratio to the distribution of ratios from genes matched in size (+-5%) to the gene in question (where the distributions from the random genes can be seen as boxplots). The white line indicates the mean; the box represents 1st and 3rd quartile and the white whiskers in the box show the 95% confidence interval. The diamond represents the ratio of the gene indicated under the box. Adj. P values for this analysis for AFF2; E2F4; MY07A; PIK3CB; and RUNX2 are, 1; 1; 0.046, 1; and 1; respectively.
Supplementary Figure 11 Overlap between three network-based methods
To compare gene predictions from several approaches, we ran HotNet2 and Muffin with the same input data as the NetSig pan cancer analysis (InWeb3 and MutSig data from 21 cancer subtypes). We defined candidate genes from HotNet 2 as all genes present in the predicted networks. We defined candidate genes from Muffinn by using a probability cut off of 0.5, as recommended by the authors. Genes shared between the methods are NetSig–HotNet2: ERBB3, RASA1 and STK11; for HotNet2–Muffinn: EGFR, SMAD4, CREBBP, EP300, TP53 and all methods predicted PIK3CA and PIK3R1.
Supplementary Figure 12 NetSig is independent of mutation frequency and identifies more low frequency cancer genes
a) A box plot of the NetSig (red) and MutSig suite (blue) P values (x-axis) versus mutation frequency distributions (y-axis). Boxes represent median first and third quartile of the frequency distribution for a given p value bin (NetSig values are permutation-based which limits us to deriving P values >=1.0e-6). In contrast to the MutSig suite, NetSig P values are not correlated with mutation frequencies. b) The proportion of all genes in the genome mutated at high, intermediate and low frequencies are shown in columns 1-3 (all genes in the genome), columns 4-6 (all significant MutSig genes using the pan-cancer data), columns 7-9 (all significant NetSig genes using the NetSig data), 10-12 (the Cancer5000 set), 13-16 (the NetSig set).
Supplementary Figure 13 Using NetSig with different functional genomics networks
To test the general applicability of our NetSig approach, and to investigate if candidate cancer genes could be robustly predicted in a range of different functional genomics networks using the statistical framework we have developed, we repeated our analysis in gene networks based on mRNA coexpression (GEO), gene coevolution profiles (CLIME), cancer synthetic lethality relationships (AchillesNet), and cell perturbation profiles (LINCS). While we observe the strongest signal in the protein-protein interaction network data from InWeb (Main text Figure 1), three of four other networks (when analyzed using NetSig) can classify known cancer genes (excluding the network based on coevolution profiles). The five Tiers of genes are defined in Supplementary Figure 1).
Supplementary Figure 14 QQ plots for different functional genomics networks
We generated qq-plots for the additional networks tested in Supplementary Figure 12. The average genomic inflation factor (lambda) is 1.14. Since these networks are based on genome-scale transcriptional datasets ‘knowledge contamination’ cannot be a factor here and the genomic inflation factor is most likely due to the polygenic nature of cancers.
Supplementary Figure 15 QQ plots when applying NetSig to regulatory networks
To corroborate the results from Supplementary Figure 13, we also ran NetSig on a collection of regulatory networks from Marbach et al. 2016. The average genomic inflation factor (lambda) is 1.11. Since these networks are based on genome-scale transcriptional data ‘knowledge contamination’ cannot be a factor here and the genomic inflation factor is most likely due to the polygenic nature of cancers.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Notes 1–10 (PDF 2659 kb)
Life Sciences Reporting Summary
Life Sciences Reporting Summary (PDF 137 kb)
Supplementary Table 1
Genes in the Tiers 1-5 used for benchmarking (XLSX 45 kb)
Supplementary Table 2
Gene-specific NetSig scores (XLSX 37 kb)
Supplementary Table 3
Literature review of NetSig5000 genes (XLSX 29 kb)
Supplementary Table 4
NetSig candidates tested experimentally (XLSX 42 kb)
Supplementary Table 5
80 barcoded cDNA constructs corresponding to 79 activating alleles of 25 known oncogenes (XLSX 33 kb)
Supplementary Table 6
80 Random genes tested experimentally (XLSX 29 kb)
Supplementary Table 7
Details on sensitivity and specificity calculations for genes (XLSX 40 kb)
Supplementary Table 8
Datasets of patients with unknown driver mutations (XLSX 25 kb)
Supplementary Table 9
Mutation rates and patterns in patients with no known driver mutations (XLSX 41 kb)
Supplementary Table 10
Candidate genes for pan cancer analysis from NetSig, Hotnet 2 and Muffinn (XLSX 34 kb)
Supplementary Software
Scripts and data to reproduce the tumorigenesis assay (ZIP 4019 kb)
Rights and permissions
About this article
Cite this article
Horn, H., Lawrence, M., Chouinard, C. et al. NetSig: network-based discovery from cancer genomes. Nat Methods 15, 61–66 (2018). https://doi.org/10.1038/nmeth.4514
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4514
This article is cited by
-
On the complexity of quantum link prediction in complex networks
Scientific Reports (2024)
-
Unraveling the Drivers of Tumorigenesis in the Context of Evolution: Theoretical Models and Bioinformatics Tools
Journal of Molecular Evolution (2023)
-
Tailored graphical lasso for data integration in gene network reconstruction
BMC Bioinformatics (2021)
-
Coexpression network architecture reveals the brain-wide and multiregional basis of disease susceptibility
Nature Neuroscience (2021)
-
Cancer gene mutation frequencies for the U.S. population
Nature Communications (2021)