Abstract
Single-molecule FISH (smFISH) has been the gold standard for quantifying individual transcript abundances. Here, we scale up multiplexed smFISH to the transcriptome level and profile 10,212 different mRNAs from mouse fibroblast and embryonic stem cells. This method, called RNA sequential probing of targets (SPOTs), provides an accurate, flexible, and low-cost alternative to sequencing for profiling transcriptomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Nat. Methods 5, 621–628 (2008).
Nagalakshmi, U. et al. Science 320, 1344–1349 (2008).
Raj, A., Peskin, C.S., Tranchina, D., Vargas, D.Y. & Tyagi, S. PLoS Biol. 4, e309 (2006).
Femino, A.M., Fay, F.S., Fogarty, K. & Singer, R.H. Science 280, 585–590 (1998).
Lubeck, E., Coskun, A.F., Zhiyentayev, T., Ahmad, M. & Cai, L. Nat. Methods 11, 360–361 (2014).
Shah, S., Lubeck, E., Zhou, W. & Cai, L. Neuron 92, 342–357 (2016).
Takei, Y., Shah, S., Harvey, S., Qi, L.S. & Cai, L. Biophys. J. 112, 1773–1776 (2017).
Frieda, K.L. et al. Nature 541, 107–111 (2017).
Lubeck, E. & Cai, L. Nat. Methods 9, 743–748 (2012).
Chen, F., Tillberg, P.W. & Boyden, E.S. Science 347, 543–548 (2015).
Beliveau, B.J. et al. Proc. Natl. Acad. Sci. USA 109, 21301–21306 (2012).
Daniel, S.G., Westling, M.E., Moss, M.S. & Kanagy, B.D. Biotechniques 24, 484–489 (1998).
Moffitt, J.R. et al. Proc. Natl. Acad. Sci. USA 113, 11046–11051 (2016).
Singer, Z.S. et al. Mol. Cell 55, 319–331 (2014).
Geiss, G.K. et al. Nat. Biotechnol. 26, 317–325 (2008).
Choi, H.M., Beck, V.A. & Pierce, N.A. ACS Nano 8, 4284–4294 (2014).
Bose, S. et al. Genome Biol. 16, 120 (2015).
Cao, J. et al. Science 357, 661–667 (2017).
Fulton, D.L. et al. Genome Biol. 10, R29 (2009).
Donner, Y., Feng, T., Benoist, C. & Koller, D. Nat. Methods 9, 1120–1125 (2012).
Eng, C.-H.L., Shah, S., Thomassie, L.J. & Cai, L. Protoc. Exch. https://doi.org/10.1038/protex.2017.131 (2017).
Engreitz, J.M. et al. Science 341, 1237973 (2013).
Antebi, Y.E. et al. Cell 170, 1184–1196.e24 (2017).
Acknowledgements
We thank P. Sims, A. Raj, B. Wold, S. Linnarson, and R. Sandberg for discussions. We thank Y. Zhang for help with automated fluidics system, N. Koulena for helping in probe generation, and E. Lubeck for help at the early stage of the project. This work was supported by grants from US National Institutes of Health (NIHRNA-1-NIHP.HD075605 and RNASUPP-1-NIHP.HD075605).
Author information
Authors and Affiliations
Contributions
C.-H.L.E. and L.C. conceived and designed experiments. C.-H.L.E. performed all the experiments. S.S. wrote the scripts for image analysis. C.-H.L.E. performed image analysis with the guidance of S.S. J.T. wrote the script for probe generations. C.-H.L.E. and L.C. performed data analysis. All authors contributed in writing the manuscripts.
Corresponding author
Ethics declarations
Competing interests
L.C., C.-H.L.E., and S.S have filed a patent on RNA SPOTs and its barcoding scheme.
Integrated supplementary information
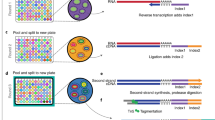
Supplementary Figure 1 RNA SPOTs hybridization and barcoding scheme.
To implement transcriptome RNA SPOTs, 5 rounds of barcoding are needed to generate >20,000 different unique error-tolerant barcodes using a 12-base coding scheme to code for the transcriptome. A round of barcoding involves 4 serial hybridizations, each of which uses three unique secondary readout probes fluorescently labeled to Alexa 647, Alexa 594, and Cy3b dyes. The images from each 4 rounds of serial hybridizations are collapsed to form each 12-pseudocolor composite image which are aligned to decode for the barcoded RNA species.
Supplementary Figure 2 Fluorescent switching through cleavage of disulfide conjugate dye on readout probes is highly efficient.
(a) 20 rounds of hybridization are accomplished by extinguishing fluorescent signals through reduction of disulfide conjugated dye to readout probes using TCEP, followed by re-hybridization of next unique secondary readout probes. (b) Both priming regions (grey in the probe schematic) used in synthesizing gene specific primary probes are also used as a registration marker through the hybridization of Alexa 488 conjugated readout probes. Majority of the fluorescent spots stay even after 20 rounds of hybridizations. The amide bond between the Alexa 488 dye (shown in yellow) and primer readout probes used as a registration marker is not affected by TCEP. (Scale bars: 2μm.) (c) The fluorescent signals in each channel after treatment of 50mM of TCEP for 5 minutes at room temperature is reduced to minimal to none. (Scale bars: 5μm.)
Supplementary Figure 3 Raw images of 20 rounds of fluorescent switching in channel 647.
Bright dots are the real targets while dim dots are due to nonspecific binding. The switching between each round of hybridization is complete, with minimal retention of fluorescent signals from previous round. (Scale bars: 2μm.)
Supplementary Figure 4 Assessment of primary probes non-specific binding.
(a) Raw images of 532 channel with the presence of mRNA on coverslips through LNA poly(d)T capturing. (b) No bright fluorescent signals are observed in the absence of mRNA on coverslips as a control. The left image has the same contrast as (a) while the right image contrast has been increased 4.5 fold to illustrate better the non-specific fluorescent signals. (c) Quantitative measurement of fluorescent intensity in channel 647 with and without the presence of mRNA. A threshold can be set to distinguish between the two populations to identify the real signals. (d) & (e) same as (c) but for channel 594 and channel 532.
Supplementary Figure 5 RNA SPOTs at lower depth.
(a) Correlation between RNA-seq FPKM and RNA SPOTs SPM from another replicate is high when a total of 376,781 spots are counted. SPM, spots per million; FPKM, fragments per kilobase per million reads. (b) High reproducibility of RNA SPOTs between the two replicates in profiling ES-E14 cell gene expression (n1=376,781 spots, n2=1,688,747 spots).
Supplementary Figure 6 Linear plots for Figure 2.
(a) Correlation between RNA SPOTs and RNA-Seq for NIH/3T3 cells. (b) Zoomed-in boxed region in (a). (c) Reproducibility between two SPOTs replicates. The dashed line corresponds to the y = x line. (d) Zoomed-in boxed region in (c). (e) Correlation between RNA SPOTs and RNA-Seq for ES-E14 cells. (f) Zoomed-in boxed region in (e).
Supplementary Figure 7 RNA SPOTs has little bias in GC content and transcript length.
(a) Hexbin plot of NIH/3T3 mean SPM (n=2) shows no obvious trend with transcript GC content. (b) Transcript length does not bias RNA SPOTs detection. (c) Same as (a) but for genes with < 1 FPKM. (d) same as (b) but for genes with < 1 FPKM. (e) Boxplots of different groups of genes with different expression levels against transcript GC content. n = 1360, 2323, 1473, 2645 for each group from left to right. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. (f) Boxplot of SPM against transcript length. (g) Replicate plot of SPOTs as shown in Fig 2b. (h) Simulated replicates with Poisson noise. The total number of simulated SPOTs (n1=447,094, n2=448,249) was set to match the experimental replicates. (i). SPOTs data from two sets of field of views (FOVs) from the E14 experiment 3, x-axis contains 25 FOVs (n1=269,459 spots), y-axis contains another 25 FOVs (n2=295,403 spots).
Supplementary Figure 8 smFISH measurement in single cells correlates with RNA SPOTs measurement in NIH/3T3 cells.
(a) Raw images of the 7 genes measured by smFISH in NIH/3T3 cells. (Scale bars: 5μm.) (b) The averaged RNA smFISH counts agrees with RNA SPOTs SPM (spots per million) with a Pearson correlation coefficient of 0.88, indicating RNA SPOTs quantitation is accurate. Error bars represents the standard error of the mean (SEM) across different single cells.
Supplementary Figure 9 mRNA can be immobilized by polyacrylamide hydrogel on a bind-silane treated coverslips.
(a) mRNA is trapped in the hydrogel mesh once acrylamide and bis-acrylamide monomers crosslink completely on the coverslip. (b) smFISH detection of ACTB once the total RNA is captured on a coverslip through LNA poly(d)T capturing (left) or polyacrylamide hydrogel (right). Negative control (channel 488) shows that the fluorescent signals are not coming from nonspecific sources. (Scale bars: 5μm.)
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Table 5 (PDF 1852 kb)
Life Sciences Reporting Summary
Life Sciences Reporting Summary (PDF 129 kb)
Supplementary Protocol
RNA SPOTs (Sequential Probing of Targets) (PDF 304 kb)
Supplementary Table 1
Primary probe sequences for 10,212 genes (XLSX 7554 kb)
Supplementary Table 2
12-base code book for 10,212 genes (XLSX 348 kb)
Supplementary Table 3
Readout probes sequences (XLSX 12 kb)
Supplementary Table 4
Spots per million in NIH/3T3 and ES-E14 cells (XLSX 612 kb)
Supplementary Table 6
Differential gene expression between mouse NIH/3T3 and ES-E14 cells (CSV 9 kb)
Supplementary Software
Analysis code for RNA SPOTs (ZIP 4 kb)
Rights and permissions
About this article
Cite this article
Eng, CH., Shah, S., Thomassie, J. et al. Profiling the transcriptome with RNA SPOTs. Nat Methods 14, 1153–1155 (2017). https://doi.org/10.1038/nmeth.4500
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4500
This article is cited by
-
scGIST: gene panel design for spatial transcriptomics with prioritized gene sets
Genome Biology (2024)
-
Single-cell analysis reveals the stromal dynamics and tumor-specific characteristics in the microenvironment of ovarian cancer
Communications Biology (2024)
-
Spatial profiling of microbial communities by sequential FISH with error-robust encoding
Nature Communications (2023)
-
HT-smFISH: a cost-effective and flexible workflow for high-throughput single-molecule RNA imaging
Nature Protocols (2023)
-
De novo reconstruction of cell interaction landscapes from single-cell spatial transcriptome data with DeepLinc
Genome Biology (2022)