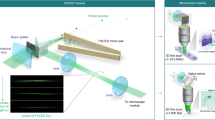
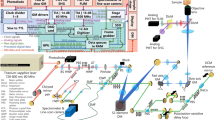
Abstract
Image scanning microscopy (ISM) doubles the resolution of a conventional confocal microscope for super-resolution imaging. Here, we describe an all-optical ISM design based on rescanning microscopy for two-photon-excited fluorescence and second-harmonic generation that allows straightforward implementation into existing microscopes. The design offers improved sensitivity and high frame rates relative to those of existing systems. We demonstrate its utility using fixed and living specimens as well as collagen hydrogels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Diaspro, A. et al. Biomed. Eng. Online 5, 36 (2006).
Hoover, E.E. & Squier, J.A. Nat. Photonics 7, 93–101 (2013).
Brown, E. et al. Nat. Med. 9, 796–800 (2003).
Campagnola, P. Anal. Chem. 83, 3224–3231 (2011).
Chen, X., Nadiarynkh, O., Plotnikov, S. & Campagnola, P.J. Nat. Protoc. 7, 654–669 (2012).
Müller, C.B. & Enderlein, J. Phys. Rev. Lett. 104, 198101 (2010).
Gustafsson, M.G.L. J. Microsc. 198, 82–87 (2000).
York, A.G. et al. Nat. Methods 9, 749–754 (2012).
Gustafsson, M.G.L. Proc. Natl. Acad. Sci. USA 102, 13081–13086 (2005).
Fiolka, R., Shao, L., Rego, E.H., Davidson, M.W. & Gustafsson, M.G.L. Proc. Natl. Acad. Sci. USA 109, 5311–5315 (2012).
Sheppard, C.J.R. Optik (Stuttg.) 80, 53–54 (1988).
Schulz, O. et al. Proc. Natl. Acad. Sci. USA 110, 21000–21005 (2013).
De Luca, G.M.R.R. et al. Biomed. Opt. Express 4, 2644–2656 (2013).
Roth, S., Sheppard, C.J.R., Wicker, K. & Heintzmann, R. Opt. Nanoscopy 2, 5 (2013).
Azuma, T. & Kei, T. Opt. Express 23, 15003–15011 (2015).
York, A.G. et al. Nat. Methods 10, 1122–1126 (2013).
Roider, C., Ritsch-Marte, M. & Jesacher, A. Opt. Lett. 41, 3825–3828 (2016).
Ingaramo, M. et al. Proc. Natl. Acad. Sci. USA 111, 5254–5259 (2014).
Winter, P.W. et al. Optica 1, 181–191 (2014).
Zipfel, W.R., Williams, R.M. & Webb, W.W. Nat. Biotechnol. 21, 1369–1377 (2003).
Roth, S., Sheppard, C.J.R. & Heintzmann, R. Opt. Lett. 41, 2109–2112 (2016).
Tinevez, J.-Y. Methods Enzymol. 506, 291–309 (2012).
Donnert, G. et al. Proc. Natl. Acad. Sci. USA 103, 11440–11445 (2006).
Ouzounov, D.G. et al. Nat. Methods 14, 388–390 (2017).
Hortn, N.G. et al. Nat. Photonics 7, 205–209 (2013).
Staunton, J.R., Doss, B.L., Lindsay, S. & Ros, R. Sci. Rep. 6, 19686 (2016).
Clarkson, M. & Saint, R. DNA Cell Biol. 18, 457–462 (1999).
Kanesaki, T., Edwards, C.M., Schwarz, U.S. & Grosshans, J. Integr. Biol. 3, 1112–1119 (2011).
Acknowledgements
Part of this work (J.E., I.G.) was supported by the Cluster of Excellence and DFG Research Center Nanoscale Microscopy and Molecular Physiology of the Brain. R.R. was supported by the NSF grant 1510700. We thank F. Rehfeldt (3rd Institute of Physics-Biophysics, University of Göttingen, Göttingen, Germany) for the hMSC samples, and J. Faust for help with the Arivis image processing.
Author information
Authors and Affiliations
Contributions
I.G. developed the setup; M.S. and I.G. built the instrument and performed fluorescence measurements. R.R. prepared the collagen samples and measured the SHG images. R.P. and J.G. provided the Drosophila embryos. J.E. conceived and directed the work. All authors discussed the results. I.G., R.R., and J.E. wrote the paper and reviewed and edited the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note 1 (PDF 2087 kb)
SHG confocal
SHG signal of a collagen I gel as recorded by confocal microscopy. (MOV 19677 kb)
SHG ISM
SHG signal of a collagen I gel as recorded by image scanning microscopy. (MOV 19684 kb)
Diffusing beads
This video shows fluorescent beads (d = 100 nm) diffusing freely in water. The recording frame rate is 20 fps. The acquisition speed is sufficient to track individual beads for several frames until they diffuse out of focus. (MOV 4866 kb)
D. melanogaster embryo development 1
Time-series of 3D stacks of living Drosophila embryos. Sections are taken around the center height of the embryo and the spacing of the z-planes is 3 μm.Frame interval is 60 s. (MOV 695 kb)
D. melanogaster embryo development 2
Time-series of 3D stacks of living Drosophila embryos. Sections are taken around the center height of the embryo and the spacing of the z-planes is 3 μm.Frame interval is 60 s. (MOV 873 kb)
D. melanogaster embryo development 3
Time-series of 3D stacks of living Drosophila embryos. Sections are taken around the center height of the embryo and the spacing of the z-planes is 3 μm.Frame interval is 60 s. (MOV 1115 kb)
D. melanogaster embryo development 4
Time-series of 3D stacks of living Drosophila embryos. Sections are taken around the center height of the embryo and the spacing of the z-planes is 3 μm.Frame interval is 120 s. (MOV 755 kb)
Source data
Rights and permissions
About this article
Cite this article
Gregor, I., Spiecker, M., Petrovsky, R. et al. Rapid nonlinear image scanning microscopy. Nat Methods 14, 1087–1089 (2017). https://doi.org/10.1038/nmeth.4467
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4467
This article is cited by
-
Signal improved ultra-fast light-sheet microscope for large tissue imaging
Communications Engineering (2024)
-
Superresolution structured illumination microscopy reconstruction algorithms: a review
Light: Science & Applications (2023)
-
Nondestructive inspection of surface nanostructuring using label-free optical super-resolution imaging
Scientific Reports (2023)
-
Video-rate multi-color structured illumination microscopy with simultaneous real-time reconstruction
Nature Communications (2019)
-
Super-resolution enhancement by quantum image scanning microscopy
Nature Photonics (2019)