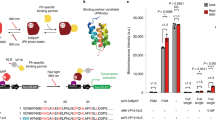
Abstract
Our improved CRISPR–Cas9-based photoactivatable transcription systems, CPTS2.0 and Split-CPTS2.0, enable high blue-light-inducible activation of endogenous target genes in various human cell lines. We achieved reversible activation of target genes with CPTS2.0 and induced neuronal differentiation in induced pluripotent stem cells (iPSCs) by upregulating NEUROD1 with Split-CPTS2.0.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dominguez, A.A., Lim, W.A. & Qi, L.S. Nat. Rev. Mol. Cell Biol. 17, 5–15 (2016).
Nihongaki, Y., Yamamoto, S., Kawano, F., Suzuki, H. & Sato, M. Chem. Biol. 22, 169–174 (2015).
Polstein, L.R. & Gersbach, C.A. Nat. Chem. Biol. 11, 198–200 (2015).
Nihongaki, Y., Kawano, F., Nakajima, T. & Sato, M. Nat. Biotechnol. 33, 755–760 (2015).
Kawano, F., Suzuki, H., Furuya, A. & Sato, M. Nat. Commun. 6, 6256 (2015).
Nihongaki, Y., Suzuki, H., Kawano, F. & Sato, M. ACS Chem. Biol. 9, 617–621 (2014).
Konermann, S. et al. Nature 517, 583–588 (2015).
Tanenbaum, M.E., Gilbert, L.A., Qi, L.S., Weissman, J.S. & Vale, R.D. Cell 159, 635–646 (2014).
Chavez, A. et al. Nat. Methods 12, 326–328 (2015).
Zetsche, B., Volz, S.E. & Zhang, F. Nat. Biotechnol. 33, 139–142 (2015).
Maeder, M.L. et al. Nat. Methods 10, 977–979 (2013).
Perez-Pinera, P. et al. Nat. Methods 10, 973–976 (2013).
Kennedy, M.J. et al. Nat. Methods 7, 973–975 (2010).
Zhang, Y. et al. Neuron 78, 785–798 (2013).
Morita, S. et al. Nat. Biotechnol. 34, 1060–1065 (2016).
Xu, X. et al. Cell Discov. 2, 16009 (2016).
Hess, G.T. et al. Nat. Methods 13, 1036–1042 (2016).
Miyamoto, T. et al. Nat. Chem. Biol. 8, 465–470 (2012).
DeRose, R., Miyamoto, T. & Inoue, T. Pflugers Arch. 465, 409–417 (2013).
Levskaya, A., Weiner, O.D., Lim, W.A. & Voigt, C.A. Nature 461, 997–1001 (2009).
Acknowledgements
This work was supported by the Japan Science and Technology Agency (START and CREST grant JPMJCR1653 to M.S.) and the Japan Society for the Promotion of Science (JSPS) (grants to M.S.; JSPS Research Fellowships for Young Scientists to Y.N. (15J05897) and Y.F. (15J08446)).
Author information
Authors and Affiliations
Contributions
Y.N. and M.S. conceived the project. Y.N., Y.F., T.O., S.H., K.Y. and M.S. designed experiments. Y.N., Y.F., T.O. and S.H. performed experiments. Y.N. analyzed data. Y.N. and M.S. wrote the manuscript. All authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Schematics of SAM, dCas9-SunTag and dCas9-VPR.
(a) Schematics of SAM. SAM consists of two chimeric proteins, dCas9 fused with VP64 and MS2-coat protein fused with p65 and HSF1 activator domains (MS2-NLS-p65-HSF1), and an sgRNA having MS2 aptamers in stem loops, known as sgRNA 2.0. MS2 aptamers in sgRNA recruits MS2-NLS-p65-HSF1, so this system can simultaneously recruit VP64, p65 and HSF1. (b) Schematics of dCas9-Suntag. dCas9-Suntag consists of two chimeric proteins, dCas9 fused with 10 copies of a GCN4 peptide and single-chain variable fragment nanobody (scFv) for GCN4 fused with VP64 activator domains (scFv-VP64), and an sgRNA. Because scFv-VP64 strongly binds to GCN4 peptides, this system can recruit multiple VP64 into Cas9-targeted locus. (c) Schematics of dCas9-VPR. VPR is a potent tripartite activator consisting of VP64, p65AD and the Epstein-Barr virus R transactivator Rta.
Supplementary Figure 2 Design and characterization of padCas9-SAM, padCas9-SunTag and padCas9-VPR.
(a) Schematics of padCas9-SAM. padCas9-SAM consists of three chimeric proteins, N-terminal fragment of dCas9 (residues 2–713, named dCas9N) fused with pMag (NLS-dCas9N-pMag), C-terminal fragment of dCas9 (residues 714–1368, named dCas9c) fused with VP64 and nMag (nMag-dCas9c-NLS-VP64) and MS2-coat protein fused with p65 and HSF1 activator domains (MS2-NLS-p65-HSF1), and an sgRNA having MS2 aptamers in stem loops, known as sgRNA 2.0. MS2 aptamers in sgRNA recruits MS2-NLS-p65-HSF1, so this system can simultaneously recruit VP64, p65 and HSF1. (b) Schematics of padCas9-Suntag. padCas9-Suntag consists of three chimeric proteins, NLS-dCas9N-pMag, C-terminal fragment of dCas9 fused with nMag and 10 copies of a GCN4 peptide (nMag-dCas9c -NLS-10xGCN4), and single-chain variable fragment nanobody (scFv) for GCN4 fused with VP64 activator domains (scFv-VP64), and an sgRNA. Because scFv-VP64 strongly binds to GCN4 peptides, this system can recruit multiple VP64 into targeted locus by light. (c) Schematics of padCas9-VPR. VPR is a potent tripartite activator consisting of VP64, p65AD and the Epstein-Barr virus R transactivator Rta. padCas9-VPR consists of two chimeric proteins, NLS-dCas9N-pMag and C-terminal fragment of dCas9 fused with nMag and VPR (nMag-dCas9c-NLS-VPR), and an sgRNA. (d) Light-induced endogenous ASCL1 activation in HEK293T cells with padCas9-Suntag, padCas9-VPR and padCas9-SAM with a single sgRNA targeting the promoter region of ASCL1. Data are expressed as relative mRNA amount to the negative control transfected with empty vector in the dark and represented as mean ± s.e.m. (n=3 from three individual experiments). D, Dark; L, Light.
Supplementary Figure 3 Optimization of padCas9-SAM.
(a) Optimizing subcellular localization of padCas9 fragments and replacing nMag photoactivatable dimerization domain. The nuclear localization signal (NLS) of NLS-dCas9N-pMag was removed and two copies of nuclear export signal (NES) were fused with dCas9N-pMag (NES-dCas9N-pMag-NES). We also replaced nMag of nMag-dCas9C-NLS-VP64 with nMagHigh1 for enhancing dimerization efficiency (nMagHigh1-dCas9C-NLS-VP64). (b) Screening of MS2 effectors that have different designs. About MS2-coat protein, we tested MS2(N55K) mutant, which is originally used in SAM, and non-aggregating MS2 delta FG mutant. We also tested whether the number and position of NLS effects induction potency. (c) Gene activation efficiencies of several MS2 effectors with NES-dCas9N-pMag-NES and nMagHigh1-dCas9C-NLS-VP64. sgRNA 2.0 targeting human ASCL1 is used. Data are expressed as relative ASCL1 mRNA amount to the negative control transfected with empty vector in the dark and represented as mean ± s.e.m. (n=4 from two individual experiments with two cell culture replicates). D, Dark; L, Light.
Supplementary Figure 4 Optogenetic activation of various genes with Split-CPTS2.0.
Light-induced upregulation of ASCL1 (a), IL1R2 (b), NEUROD1 (c), IL1RN (d) and MYOD1 (e) in HEK293T cells using Split-CPTS2.0, CPTS, LACE, dCas9-VP64 and SAM with a single sgRNA targeting each gene. Data are expressed as relative mRNA amount to the negative control transfected with empty vector in the dark and represented as mean ± s.e.m. ((a–d) n=4 from two individual experiments with two cell culture replicates. (e) n=6 from three individual experiments with two cell culture replicates.). D, Dark; L, Light; ND, Not determined. Welch’s two-tailed t test was performed. *p < 0.05, ** < 0.01, ***p < 0.005 versus the sample in the dark.
Supplementary Figure 5 Split-CPTS2.0 can induce potent gene activation in various human cells.
Light-induced ASCL1 upregulation in HeLa cells (a) and human fetal fibroblasts (b) using Split-CPTS2.0, CPTS, LACE, dCas9-VP64 and SAM with a single sgRNA. Data are expressed as relative ASCL1 mRNA amount to the negative control transfected with empty vector or EGFP in the dark and represented as mean ± s.e.m. ((a) n=4 from two individual experiments with two cell culture replicates. (b) n=2 from two individual experiments). D, Dark; L, Light.
Supplementary Figure 6 Split-CPTS2.0-mediated optogenetic gene activation with multiple sgRNAs.
Light-induced upregulation of MYOD1 (a) and IL1RN (b) in HEK293T cells using Split-CPTS2.0 and CPTS with a single sgRNA and quadruple sgRNAs targeting MYOD1 and IL1RN, respectively. Data are expressed as relative mRNA amount to the negative control transfected with empty vector in the dark and represented as mean ± s.e.m. (n=6 from three individual experiments with two cell culture replicates). D, Dark; L, Light. Welch’s two-tailed t test was performed. *p < 0.05, ** < 0.01, ***p < 0.005 versus the sample in the dark.
Supplementary Figure 7 Spatial surrogate reporter activation by Split-CPTS2.0.
(a) Schematic of GAL4-UAS mCherry reporter. (b) Light-induced mCherry reporter activation of Split-CPTS2.0. HEK293T cells were transfected with Split-CPTS2.0, GAL4-UAS mCherry reporter and sgRNA targeting the reporter. Twenty hours post transfection, samples were incubated under dark state, global blue light illumination for 24 h. Scale bar = 2 mm. (c) Spatial gene activation by Split-CPTS2.0. Slit-patterned mCherry expression in HEK293T cells illuminated by blue light with a spatial pattern using a photomask. EGFP is used as transfection marker. The width of slit is 2.6 mm. Close-up view within white square region is also shown. Scale bar, 2 mm (zoom-out view) and 0.5 mm (close-up view), respectively. (d) Line scan intensity profile of EGFP (turquoise) and mCherry (magenta) on dashed line in c.
Supplementary Figure 8 CPTS2.0-mediated optogenetic endogenous gene activation.
(a,b) Light-induced upregulation of ASCL1 (a) and MYOD1 (b) in HEK293T cells. Used dCas9-activators and the number of sgRNAs are indicated. (c,d) Light-induced IL1RN upregulation in HeLa cells (c) and human induced pluripotent stem cells (d) using CPTS, CPTS2.0 and Split-CPTS2.0 with a single sgRNA targeting IL1RN gene. (e) Reversible and repeatable activation of endogenous ASCL1 with CPTS2.0. Data are expressed as relative mRNA amount to the negative control transfected with empty vector in the dark and represented as mean ± s.e.m. (n=4 from two individual experiments with two cell culture replicates). D, Dark; L, Light. Welch’s two-tailed t test was performed. *p < 0.05, ** < 0.01, ***p < 0.005 versus the sample in the dark.
Supplementary Figure 9 Comparison of light-induced MYOD1 and IL1RN activation efficiency among various sgRNAs targeting different sites in the promoter regions.
Light-induced upregulation of MYOD1 (a) and IL1R2 (b) in HEK293T cells using Split-CPTS2.0, CPTS2.0, CPTS and SAM with an indicated single sgRNA. Maps show the sgRNA targeting sites in each human locus. Data are expressed as relative mRNA amount to the negative control transfected with empty vector in the dark and represented as mean ± s.e.m. (n=4 from two individual experiments with two cell culture replicates).
Supplementary Figure 10 DNA-targeting specificity analysis of Split-CPTS2.0 and CPTS2.0.
(a) Luciferase reporter is constructed of Cas9-binding sites, minimal CMV promoter and luciferase gene. (b) DNA targeting specificity analysis of Split-CPTS2.0, CPTS2.0, dCas9-VP64 and SAM using luciferase reporter plasmid activation assay. The positions of mutations in each sgRNA are indicated in red. Values are normalized to positive control with perfectly matched sgRNAs. Data are shown as the mean ± s.e.m. (n=6 from two individual experiments with three cell culture replicates).
Supplementary Figure 11 NEUROD1 activation kinetics in human iPSCs with dCas9 activators.
(a) Light-induced NEUROD1 upregulation in human iPSCs using indicated dCas9 activators with indicated sgRNAs at day 4. The negative sgRNA targets unrelated DNA sequence. (b) Time-course of NEUROD1 activation in human iPSCs using indicated dCas9 activators. Data are expressed as relative mRNA amount to the negative control transfected with EGFP vector in the dark and are shown as the mean ± s.e.m. (n=3 three cell culture replicates). D, Dark; L, Light; ND, Not determined. Welch’s two-tailed t test was performed. ***p < 0.005 versus the sample in the dark.
Supplementary Figure 12 Representative images of Split-CPTS2.0-induced neurons.
Immunofluorescence images of Split-CPTS2.0-induced neurons from human iPSCs (454E2) for beta III tubulin (magenta) and DAPI (blue). Immunofluorescence images were taken 4 days after blue light stimulation or dark incubation. Scale bar = 50 μm.
Supplementary Figure 13 CPTS could not induce neuronal differentiation of human iPSCs.
Immunofluorescence images of human iPSCs for beta III tubulin (magenta), DAPI (blue) and EGFP (Green) after 96 h blue light illumination or incubation in the dark. Transfected plasmids are indicated. Images are representative from three individual experiments. Scale Bar = 50 μm.
Supplementary Figure 14 Split-CPTS2.0-mediated neuronal differentiation of human iPSCs (201B7).
(a) Immunofluorescence images of 201B7-derived neurons induced by split-CPTS2.0 targeting NEUROD1. Cells were immunostained after 4 days blue light stimulation or dark incubation. (b) Immunofluorescence images of 201B7 cells expressing indicated dCas9-based photoactivatable transcription systems and sgRNAs. Cells were immunostained after 4 days blue light stimulation or dark incubation. Magenta, beta III tubulin; Blue, DAPI; Green, EGFP. Scale bar = 50 μm.
Supplementary Figure 15 CPTS2.0 induced higher NEUROD1 expression in human iPSCs than CPTS but was still insufficient to induce neuronal differentiation.
(a) Light-induced NEUROD1 upregulation in human iPSCs using indicated dCas9-activators with four sgRNAs targeting NEUROD1 at day 1. Data are expressed as relative mRNA amount to the negative control transfected with EGFP vector in the dark and represented as mean ± s.e.m. (n=3 cell culture replicates from one of two representative experiments). (b) Immunofluorescence images of human iPSCs expressing CPTS2.0 targeting NEUROD1 with four sgRNAs. Magenta, beta III tubulin; Blue, DAPI. In this experiment, no positive beta III tubulin signal was observed. Immunostaining were performed after 96 h blue light illumination or incubation in the dark. Scale Bar = 50 μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Table 1 and Supplementary Notes 1–2.
Supplementary Table 2
Target sequences of sgRNAs and oligonucleotide sequences for constructing sgRNAs.
Rights and permissions
About this article
Cite this article
Nihongaki, Y., Furuhata, Y., Otabe, T. et al. CRISPR–Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat Methods 14, 963–966 (2017). https://doi.org/10.1038/nmeth.4430
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4430