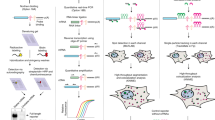
Abstract
Conversion of adenosine to inosine is a frequent type of RNA editing, but important details about the biology of this conversion remain unknown because of a lack of imaging tools. We developed inoFISH to directly visualize and quantify adenosine-to-inosine-edited transcripts in situ. We found that editing of the GRIA2, EIF2AK2, and NUP43 transcripts is uncorrelated with nuclear localization and paraspeckle association. Further, NUP43 exhibits constant editing levels between single cells, while GRIA2 editing levels vary.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Change history
20 July 2017
In the version of this article initially published online, a statement in the introductory paragraph incorrectly implied that deamination leads to defects in hematopoiesis and neurological functions. It is the lack of deamination that causes these defects. The error has been corrected in the print, PDF and HTML versions of this article.
References
Bass, B.L. & Weintraub, H. Cell 48, 607–613 (1987).
Liddicoat, B.J. et al. Science 349, 1115–1120 (2015).
Higuchi, M. et al. Nature 406, 78–81 (2000).
Kumar, M. & Carmichael, G.G. Proc. Natl. Acad. Sci. USA 94, 3542–3547 (1997).
Chen, L.-L. & Carmichael, G.G. Mol. Cell 35, 467–478 (2009).
Zhang, Z. & Carmichael, G.G. Cell 106, 465–475 (2001).
Prasanth, K.V. et al. Cell 123, 249–263 (2005).
Raj, A., van den Bogaard, P., Rifkin, S.A., van Oudenaarden, A. & Tyagi, S. Nat. Methods 5, 877–879 (2008).
Levesque, M.J., Ginart, P., Wei, Y. & Raj, A. Nat. Methods 10, 865–867 (2013).
Seeburg, P.H., Single, F., Kuner, T., Higuchi, M. & Sprengel, R. Brain Res. 907, 233–243 (2001).
Yamashita, T. et al. Neurosci. Res. 73, 42–48 (2012).
Paschen, W., Hedreen, J.C. & Ross, C.A. J. Neurochem. 63, 1596–1602 (1994).
Melcher, T. et al. Nature 379, 460–464 (1996).
O'Connell, M.A., Gerber, A. & Keller, W. J. Biol. Chem. 272, 473–478 (1997).
Yoshida, M. & Ukita, T. Biochim. Biophys. Acta 157, 455–465 (1968).
Sakurai, M., Yano, T., Kawabata, H., Ueda, H. & Suzuki, T. Nat. Chem. Biol. 6, 733–740 (2010).
Wang, I.X. et al. Cell Rep. 5, 849–860 (2013).
Shaffer, S.M. et al. Lab Chip 15, 3170–3182 (2015).
Sunwoo, H. et al. Genome Res. 19, 347–359 (2009).
Mellis, I.A. et al. Protocol Exchange DOI: http://dx.doi.org/10.1038/protex.2017.056 (2017).
Duffy, D.J. et al. Oncotarget 6, 43182–43201 (2015).
Kuhlwilm, M., Davierwala, A. & Pääbo, S. PLoS One 8, e83218 (2013).
Whitney, N.P. et al. FASEB J. 22, 2888–2900 (2008).
Cabili, M.N. et al. Genome Biol. 16, 20 (2015).
Acknowledgements
We thank K. Nishikura and M. Sakurai for helpful discussions, and we thank C. Bartman and B. Emert for comments and suggestions. I.A.M. acknowledges support from NIH/NIGMS T32GM007170 (University of Pennsylvania MSTP), NIH/NHGRI T32HG000046 and NIH/NINDS F30NS100595; S.H.R. from NIH 1F32GM120929-01A1; and A.R. from NSF CAREER Award 1350601, NIH New Innovator 1DP2OD008514, NIH/NIBIB R33 EB019767, NIH 4DN U01 HL129998, and NIH Center for Photogenomics RM1 HG007743.
Author information
Authors and Affiliations
Contributions
S.H.R., I.A.M., and A.R. conceived of the paper; S.H.R. and I.A.M. performed experiments; I.A.M., R.G., and A.R. wrote custom software; I.A.M., S.H.R., and A.R. analyzed the data; I.A.M., S.H.R., and A.R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
A.R. receives royalties and consulting income from Biosearch Technologies related to Stellaris RNA FISH.
Integrated supplementary information
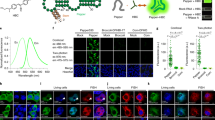
Supplementary Figure 1 Probe hybridization scheme.
(Top) Hybridization mix including all fluorescently labeled probes is added to cells. Colocalization of guide probes with detection probes determines sites of adenosine-containing or inosine-containing transcripts. (Bottom) Multiple, singly labeled probes target the transcript of interest flanking the editing site. Individual probes targeting the editing site are generated and labeled with 2 different fluorophores--these probes contain matching mask-binding regions. When the mask is bound, the region that binds to the target mRNA is shortened and a mismatch destabilizes the hybridization. The two detection probes compete for binding and the specific probe remains bound. The mask is removed by strand displacement and the full sequence hybridizes to its target.
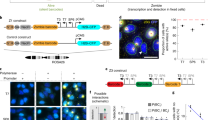
Supplementary Figure 2 Editing targets of inoFISH.
(a) GRIA2, (b) EIF2 AK2 and (c) NUP43: schematic gene representation, 5’ to 3’ regardless of strand (top), genotyping by Sanger sequencing of genomic DNA PCR around locus encoding editing site in cell line of interest (middle), and RT-PCR/Sanger verification of RNA editing in cell line of interest (bottom).
Supplementary Figure 3 GRIA2 mRNA nuclear retention analysis.
(a) Most GRIA2 transcripts are retained in the nucleus in SH-SY5Y cells. (b) GRIA2 guide probe specificity assessed by odds-evens probe set division experiment. (c) Nuclear retention of GRIA2 upon treatment with aphidicolin, a cell cycle inhibitor. (d) Western blot of GluR2 protein in SH-SY5Y cell lysate.
Supplementary Figure 4 Expanded controls for GRIA2, EIF2AK2 and NUP43 inoFISH.
InoFISH results for each replicate, including pixel-shift and dye-swap controls for GRIA2 in SH-SY5Y cells (top), EIF2AK2 in U-87 MG cells (middle) and NUP43 in U-87 MG cells (bottom).
Supplementary Figure 5 False colocalization of GRIA2 detection probes with SFPQ guide probe.
(top) Schematic representation of the experiment. Control experiment is normal GRIA2 inoFISH with GRIA2 detection probes. On the right, the guide probe has been replaced to target SFPQ with the GRIA2 detection probes. (bottom) Fraction of colocalized guides with GRIA2 (left) and SFPQ (right).
Supplementary Figure 6 Resampled mean GRIA2 editing level estimates from ADAR2 knockdown and cyanoethylation experiments.
(a) ADAR2 transcript abundance upon siRNA-mediated ADAR2 knockdown (left); GRIA2 inoFISH results in SH-SY5Y cells (n = 2) after ADAR2 knockdown and scrambled siRNA control (right). (b) Cyanoethylation reaction schematic (left) and GRIA2 inoFISH (right) results in SH-SY5Y cells (n = 3) after in situ cyanoethylation with -acrylonitrile control. (c) Resampled mean editing levels of scrambled siRNA vs ADAR2 siRNA knockdown. (d) Resampled mean editing levels of samples treated by cyanoethylation and control samples (-acrylonitrile). 10000 resampled mean estimated editing level values per boxplot.
Supplementary Figure 7 Cyanoethylation reduces number of detected inosines in situ.
inoFISH mean +/- s.e.m after cyanoethylation treatment from a minimum of 2 biological replicates (left) including pixel-shift and dye-swap controls per replicate (right) for GRIA2 in SH-SY5Y cells (top), EIF2AK2 in U87 MG cells (middle), and NUP43 in U87 MG cells (bottom).
Supplementary Figure 8 GRIA2 editing level by method.
(a) Pipeline outline for RNA-seq-based screen and GRIA2 editing level estimation. (b) Experimental pipeline and representative Sanger sequencing trace for RT-PCR/Sanger-based estimate of GRIA2 editing level. (c) Experimental pipeline and representative Sanger sequencing traces from genotyping of RT-PCR/Topo TA cloning-based estimate of GRIA2 editing level. (d) Experimental pipeline and Bioanalyzer report for RT-PCR/BbvI-digestion-based estimate of GRIA2 editing level.
Supplementary Figure 9 Comparison of inoFISH with targeted and off-target detection probes.
(top) Schematic representation of the experiment. Control experiment is normal GRIA2 inoFISH with GRIA2 detection probes. On the right, the guide probe has been rcolocalized with an adenosine detection probe and a cytosine detection probe that doesn’t target either nucleotide. (bottom) Fraction of colocalized GRIA2 guides by normal inoFISH (left) and inoFISH with false competition (right).
Supplementary Figure 10 Mean editing level estimate distributions for all three targets.
Boxplots of 10000 parametric bootstrapped samples of editing level per boxplot for GRIA2 (top), EIF2AK2 (middle), and NUP43 (bottom). Mean editing level models resampled irrespective of subcellular localization (left) and considering nuclear vs cytoplasmic localization (right).
Supplementary Figure 11 How to pick an inoFISH target.
Outline of experimental design pipeline for inoFISH, including representative results (LSG1 RT-PCR/Sanger and MYO1 C genotyping/Sanger) at critical steps in the target selection process.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Note 1. (PDF 2439 kb)
Supplementary Table 1
Sequences of oligonucleotides used. (XLSX 17 kb)
Supplementary Protocol
inoFISH Supplementary Protocol. (PDF 441 kb)
Rights and permissions
About this article
Cite this article
Mellis, I., Gupte, R., Raj, A. et al. Visualizing adenosine-to-inosine RNA editing in single mammalian cells. Nat Methods 14, 801–804 (2017). https://doi.org/10.1038/nmeth.4332
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4332
This article is cited by
-
Semi-quantitative detection of pseudouridine modifications and type I/II hypermodifications in human mRNAs using direct long-read sequencing
Nature Communications (2023)
-
Illuminating RNA biology through imaging
Nature Cell Biology (2022)
-
Achieving single nucleotide sensitivity in direct hybridization genome imaging
Nature Communications (2022)
-
Spatial epitranscriptomics reveals A-to-I editome specific to cancer stem cell microniches
Nature Communications (2022)
-
ClampFISH detects individual nucleic acid molecules using click chemistry–based amplification
Nature Biotechnology (2019)