Abstract
Studies of human T cell development require robust model systems that recapitulate the full span of thymopoiesis, from hematopoietic stem and progenitor cells (HSPCs) through to mature T cells. Existing in vitro models induce T cell commitment from human HSPCs; however, differentiation into mature CD3+TCR-αβ+ single-positive CD8+ or CD4+ cells is limited. We describe here a serum-free, artificial thymic organoid (ATO) system that supports efficient and reproducible in vitro differentiation and positive selection of conventional human T cells from all sources of HSPCs. ATO-derived T cells exhibited mature naive phenotypes, a diverse T cell receptor (TCR) repertoire and TCR-dependent function. ATOs initiated with TCR-engineered HSPCs produced T cells with antigen-specific cytotoxicity and near-complete lack of endogenous TCR Vβ expression, consistent with allelic exclusion of Vβ-encoding loci. ATOs provide a robust tool for studying human T cell differentiation and for the future development of stem-cell-based engineered T cell therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Schmitt, T.M. & Zúñiga-Pflücker, J.C. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749–756 (2002).
De Smedt, M., Hoebeke, I. & Plum, J. Human bone marrow CD34+ progenitor cells mature to T cells on OP9-DL1 stromal cell line without thymus microenvironment. Blood Cells Mol. Dis. 33, 227–232 (2004).
La Motte-Mohs, R.N., Herer, E. & Zúñiga-Pflücker, J.C. Induction of T cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood 105, 1431–1439 (2005).
de Pooter, R. & Zúñiga-Pflücker, J.C. T cell potential and development in vitro: the OP9-DL1 approach. Curr. Opin. Immunol. 19, 163–168 (2007).
Awong, G., Herer, E., La Motte-Mohs, R.N. & Zúñiga-Pflücker, J.C. Human CD8 T cells generated in vitro from hematopoietic stem cells are functionally mature. BMC Immunol. 12, 22 (2011).
Van Coppernolle, S. et al. Functionally mature CD4 and CD8 TCR-αβ cells are generated in OP9-DL1 cultures from human CD34+ hematopoietic cells. J. Immunol. 183, 4859–4870 (2009).
Hao, Q.L. et al. Human intrathymic lineage commitment is marked by differential CD7 expression: identification of CD7− lympho-myeloid thymic progenitors. Blood 111, 1318–1326 (2008).
De Smedt, M. et al. T lymphoid differentiation potential measured in vitro is higher in CD34+CD38−/lo hematopoietic stem cells from umbilical cord blood than from bone marrow and is an intrinsic property of the cells. Haematologica 96, 646–654 (2011).
Anderson, G., Jenkinson, E.J., Moore, N.C. & Owen, J.J.T. MHC class II–positive epithelium and mesenchyme cells are both required for T cell development in the thymus. Nature 362, 70–73 (1993).
Plum, J., De Smedt, M., Defresne, M.P., Leclercq, G. & Vandekerckhove, B. Human CD34+ fetal liver stem cells differentiate to T cells in a mouse thymic microenvironment. Blood 84, 1587–1593 (1994).
Poznansky, M.C. et al. Efficient generation of human T cells from a tissue-engineered thymic organoid. Nat. Biotechnol. 18, 729–734 (2000).
Chung, B. et al. Engineering the human thymic microenvironment to support thymopoiesis in vivo. Stem Cells 32, 2386–2396 (2014).
Awong, G., Motte-Mohs, R.N.L. & Zúñiga-Pflücker, J.C. In vitro human T cell development directed by notch–ligand interactions. in Hematopoietic Stem Cell Protocols. (ed. Bunting, K.D.) 135–142 (Humana Press, 2008).
Sheridan, J.M., Taoudi, S., Medvinsky, A. & Blackburn, C.C. A novel method for the generation of reaggregated organotypic cultures that permits juxtaposition of defined cell populations. Genesis 47, 346–351 (2009).
Itoh, K. et al. Reproducible establishment of hemopoietic supportive stromal cell lines from murine bone marrow. Exp. Hematol. 17, 145–153 (1989).
Brewer, G.J., Torricelli, J.R., Evege, E.K. & Price, P.J. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35, 567–576 (1993).
Huijskens, M.J.A.J. et al. Technical advance: ascorbic acid induces development of double-positive T cells from human hematopoietic stem cells in the absence of stromal cells. J. Leukoc. Biol. 96, 1165–1175 (2014).
Manning, J. et al. Vitamin C promotes maturation of T cells. Antioxid. Redox Signal. 19, 2054–2067 (2013).
Casero, D. et al. Long noncoding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 16, 1282–1291 (2015).
Awong, G. et al. Characterization in vitro and engraftment potential in vivo of human progenitor T cells generated from hematopoietic stem cells. Blood 114, 972–982 (2009).
Vanhecke, D., Leclercq, G., Plum, J. & Vandekerckhove, B. Characterization of distinct stages during the differentiation of human CD69+CD3+ thymocytes and identification of thymic emigrants. J. Immunol. 155, 1862–1872 (1995).
Res, P., Blom, B., Hori, T., Weijer, K. & Spits, H. Downregulation of CD1 marks acquisition of functional maturation of human thymocytes and defines a control point in late stages of human T cell development. J. Exp. Med. 185, 141–151 (1997).
Kimmig, S. et al. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J. Exp. Med. 195, 789–794 (2002).
Hinrichs, C.S. et al. Adoptively transferred effector cells derived from naive rather than central memory CD8+ T cells mediate superior antitumor immunity. Proc. Natl. Acad. Sci. USA 106, 17469–17474 (2009).
Restifo, N.P., Dudley, M.E. & Rosenberg, S.A. Adoptive immunotherapy or cancer: harnessing the T cell response. Nat. Rev. Immunol. 12, 269–281 (2012).
Gurka, S., Dirks, S., Photiadis, J. & Kroczek, R.A. Expression analysis of surface molecules on human thymic dendritic cells with the 10th HLDA Workshop antibody panel. Clin. Transl. Immunology 4, e47 (2015).
Martínez, V.G. et al. A discrete population of IFN-λ-expressing BDCA3hi dendritic cells is present in human thymus. Immunol. Cell Biol. 93, 673–678 (2015).
Hao, Q.L., Shah, A.J., Thiemann, F.T., Smogorzewska, E.M. & Crooks, G.M. A functional comparison of CD34+CD38− cells in cord blood and bone marrow. Blood 86, 3745–3753 (1995).
Kohn, L.A. et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat. Immunol. 13, 963–971 (2012).
Six, E.M. et al. A human postnatal lymphoid progenitor capable of circulating and seeding the thymus. J. Exp. Med. 204, 3085–3093 (2007).
Galy, A., Travis, M., Cen, D. & Chen, B. Human T, B, natural killer and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity 3, 459–473 (1995).
Gschweng, E.H. et al. HSV-sr39TK positron emission tomography and suicide gene elimination of human hematopoietic stem cells and their progeny in humanized mice. Cancer Res. 74, 5173–5183 (2014).
Snauwaert, S. et al. In vitro generation of mature, naive antigen-specific CD8+ T cells with a single T cell receptor by agonist selection. Leukemia 28, 830–841 (2014).
Giannoni, F. et al. Allelic exclusion and peripheral reconstitution by TCR transgenic T cells arising from transduced human hematopoietic stem and progenitor cells. Mol. Ther. 21, 1044–1054 (2013).
Zhao, Y. et al. Extrathymic generation of tumor-specific T cells from genetically engineered human hematopoietic stem cells via Notch signaling. Cancer Res. 67, 2425–2429 (2007).
van Lent, A.U. et al. Functional human antigen-specific T cells produced in vitro using retroviral T cell receptor transfer into hematopoietic progenitors. J. Immunol. 179, 4959–4968 (2007).
Gattinoni, L. et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J. Clin. Invest. 115, 1616–1626 (2005).
Hinrichs, C.S. & Rosenberg, S.A. Exploiting the curative potential of adoptive T cell therapy for cancer. Immunol. Rev. 257, 56–71 (2014).
Vatakis, D.N. et al. Introduction of exogenous T cell receptors into human hematopoietic progenitors results in exclusion of endogenous T cell receptor expression. Mol. Ther. 21, 1055–1063 (2013).
Stärck, L., Popp, K., Pircher, H. & Uckert, W. Immunotherapy with TCR-redirected T cells: comparison of TCR-transduced and TCR-engineered hematopoietic stem cell–derived T cells. J. Immunol. 192, 206–213 (2014).
Torikai, H. et al. A foundation for universal T cell–based immunotherapy: T cells engineered to express a CD19-specific chimeric antigen receptor and eliminate expression of endogenous TCR. Blood 119, 5697–5705 (2012).
Berdien, B., Mock, U., Atanackovic, D. & Fehse, B. TALEN-mediated editing of endogenous T cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene Ther. 21, 539–548 (2014).
Poirot, L. et al. Multiplex genome-edited T cell manufacturing platform for 'off-the-shelf' adoptive T cell immunotherapies. Cancer Res. 75, 3853–3864 (2015).
Themeli, M., Rivière, I. & Sadelain, M. New cell sources for T cell engineering and adoptive immunotherapy. Cell Stem Cell 16, 357–366 (2015).
Seet, C.S., Crooks, G.M. & Montel-Hagen, A. Artificial thymic organoid cultures: in vitro human T cell differentiation from hematopoietic stem and progenitor cells. Protoc. Exch. (http://dx.doi.org/10.1038/protex.2017.062).
Majeti, R., Park, C.Y. & Weissman, I.L. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 1, 635–645 (2007).
Robbins, P.F. et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol. 180, 6116–6131 (2008).
Johnson, L.A. et al. Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J. Immunol. 177, 6548–6559 (2006).
Giudicelli, V., Chaume, D. & Lefranc, M.-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 33, D256–D261 (2005).
Kent, W.J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Acknowledgements
We thank J. Scholes and F. Codrea at the UCLA Broad Stem Cell Research Center (BSCRC) Flow Cytometry Core for assistance with FACS sorting, R. Chan for assistance with specimen processing, C. Parekh (Children's Hospital Los Angeles) for generous assistance with thymus samples, M. Sehl (UCLA) for assistance with MPB collection, and A. Cooper (UCLA) for helpful advice and discussion. We thank I. Antoshechkin (Millard and Muriel Jacobs Genetics and Genomics Laboratory, Caltech), who developed the method for, and who assisted with, TCR sequencing analysis, A. Ribas (UCLA) for the NY-ESO-1 and MART-1 TCR constructs, J. Zuniger-Pflucker (University of Toronto) for OP9-DL1 cells, L. Coulombel for MS-5 cells and J. Chute (UCLA) for U266 cells. This work was supported by the NIH (grants R01 AG049753 (G.M.C.), 1R21AI119927 (G.M.C. and A.M.-H.), P01 HL073104 (G.M.C. and D.B.K.) and T32HL066992 (C.S.S.)), the Tower Cancer Research Foundation (C.S.S.), a UCLA BSCRC Innovation award (G.M.C. and D.B.K.) and a BSCRC Clinical Fellowship (C.S.S.). M.T.B. and D.B. are supported by Prostate Cancer Foundation Challenge Award 15CHAL02, and M.T.B. is the recipient of a Jane Coffin Childs Postdoctoral Fellowship. Core services were supported by the UCLA Jonsson Comprehensive Cancer Center shared facility (TPCL, grant 5P30CA016042), the UCLA Immunogenetics Center, the UCLA Center for AIDS Research Virology Core Lab and the UCLA AIDS Institute (grant 5P30 AI028697), and the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech.
Author information
Authors and Affiliations
Contributions
C.S.S. and A.M.-H. designed and performed experiments, analyzed data, prepared figures and co-wrote the manuscript; C.H. performed histological experiments and, with B.C., assisted with in vivo experiments; S.L. assisted with ATO analysis and T cell functional assays; K.K. assisted with ATO cultures; Y.Z. performed human specimen processing and cultured the cell lines; E.H.G. and D.B.K. provided critical reagents and conceptual advice and edited the manuscript; M.T.B. and D.B. devised the approach for, and performed, TCR repertoire sequencing analysis and provided critical reagents; and G.M.C. and A.M.-H. co-directed the project and co-wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Kite Pharma, Inc. is supporting the preclinical research of the ATO system at UCLA with G.M.C. as principal investigator. Kite Pharma, Inc. also holds an exclusive license to certain intellectual property that relates to the ATO system.
Integrated supplementary information
Supplementary Figure 1 ATOs form solid tissue-like structures.
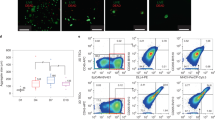
Hematoxylin and eosin staining showing tissue architecture of week 6 3D cultures generated with CB HSPCs and MS5-hDLL1 (i.e. ATO) (left), parental MS-5 cells (center), or MS5-hDLL1 cells alone (right). Magnification is 100X (top row) or 400X (bottom row).
Supplementary Figure 2 The starting number of HSPCs per ATO affects cell yield per HSPC but not the total cell output or T cell differentiation.
(a) Total cell number and yield per input HSPC in week 6 ATOs generated with varying numbers of CD34+CD3- CB HSPCs (0.3-30x103 per ATO) and a constant number of MS5-hDLL1 stromal cells (1.5x105 per ATO). Comparison is shown at far right of each graph with larger ATOs (using 30x103 HSPC and 6x105 stromal cells, at a ratio of 1:20). (b) T cell precursor and mature T cell frequencies in ATOs as described in (a). Mean and SD of triplicate ATOs are shown. Data are representative of two independent experiments.
Supplementary Figure 3 T cell differentiation in ATOs is highly reproducible and not affected by B27 lot variation, xeno-free B27 or stromal irradiation.
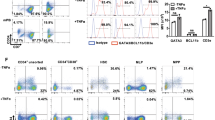
No significant effect of B27 lot variation on (a) T-lineage commitment, (b-c) T cell differentiation, or (d) total cell numbers in week 6 ATOs generated from a single CB (7.5x103 CD34+CD3- HSPCs per ATO) and cultured using 4 different lots of B27 supplement (labeled A-D). Replicate ATOs (n=2-3) are shown for each B27 lot. Substitution of standard B27 with xeno-free B27 had no significant impact on (e) T cell differentiation or (f) total cell numbers in week 6 ATOs. Irradiation of MS5-hDLL1 stromal cells with 20-80 Gy prior to ATO generation had little impact on (g-i) T cell differentiation, or (j) numbers of total cells and CD3+TCRαβ+CD8SP T cells. Mean and SD of triplicate ATOs are shown. Data are representative of two individual experiments. Flow plots in (h) show cells from CD3+TCRαβ+ gate shown in (g). (k) Harvesting cells from ATOs by mechanical disruption at 6 weeks resulted in a suspension of >99% human hematopoietic CD45+ cells (top), and <0.5% GFP+ stromal cells (bottom). Frequencies are shown for 8 independent experiments (error bars represent SD).
Supplementary Figure 4 Enhanced T cell positive selection and maturation in ATOs compared with OP9-DL1 monolayer co-cultures.
Full flow cytometry data for Fig. 2a is provided, showing T cell differentiation from three different cord blood donors (#1, #2 and #3) at (a) 4 weeks and (b) 6 weeks. ATO and OP9-DL1 cultures were started in parallel with CD34+CD3- HSPCs from the same cord blood units. Cells are gated on total CD14-CD56- cells or CD3+TCRαβ+ T cells as indicated. (c) Absolute cell numbers of T cell subsets at week 4 and 6 in OP9-DL1 co-cultures versus ATOs using the gating strategy shown in (a) and (b). Each OP9-DL1 culture was initiated with 1.5x104 CD34+CD3- CB HSPCs cells, and each ATO was initiated with 7.5x103 HSPCs from the same cord blood unit, with technical duplicate ATOs harvested and pooled at the indicated times for comparison of cell counts. Bars represent the mean and SD of three independent experiments.
Supplementary Figure 5 Enhanced positive selection in ATOs requires 3D structure and optimal cell line and culture medium.
Enhanced T cell positive selection and maturation in ATOs (defined as MS5-hDLL1 in 3D culture with RB27) compared with monolayer co-cultures. Week 6 monolayer cultures (left) were compared with 3D organoid cultures (right) and included crossover comparisons with either MS5-DL1 or OP9-DL1 cells and RB27 medium or OP9-DL1 standard medium as indicated. Standard medium for OP9-DL1 co-cultures was MEMα/20%FCS with IL-7, FLT3L, and ascorbic acid, and standard medium for ATOs was RB27 with IL-7, FLT3L, and ascorbic acid, as described in Methods. Monolayer or 3D cultures using the parental MS-5 cell line (not transduced with DLL1) are also shown as a negative control. All plots are gated on CD14-CD56- cells or (where indicated) CD3+TCRαβ+ subgates.
Supplementary Figure 6 Recapitulation of thymopoiesis and naive T cell phenotype in ATOs.
(a) Progressive differentiation of naïve CD3+TCRαβ+CD8SP and CD3+TCRαβ+CD4SP cells in ATOs between weeks 6-10. ATOs were cultured in parallel using CB HSPCs from a single donor and analyzed at the indicated weeks. Cells are gated on CD14-CD56-TCRαβ+CD3+ cells, and sequential sub-gates (CD8SP or CD4SP) are indicated above plots. The corresponding week 12 timepoint is shown in Fig. 2. (b,c) Additional markers characterizing the phenotype of week 12 ATO-derived CD3+TCRαβ+ (b) CD8SP and (c) CD4SP T cells compared with corresponding populations in the human thymus (see also Fig. 2). (d) Frequency of HLA-DR+ cells in CB ATOs compared with postnatal thymi (gated on total CD45+ cells). (e) Multiple HLA-DR+ antigen presenting cell (APC) populations are present in week 6 ATOs. Sequential gates are shown above each plot. HLA-DR+ populations include monocytes (CD14+), granulocytes (CD66b+), B cells (CD19+), HSPCs (CD34+), plasmacytoid DC (CD303+CD123+), CLEC9A+ DC (CD141+CLEC9A+), and CD1c+ DC (CD1c+CLEC9A-). Paired analysis from a postnatal thymus is shown for comparison. Data in (d) and (e) are representative of three independent experiments.
Supplementary Figure 7 Generation of T cells from multiple HSPC sources and subsets.
(a) Persistence of CD34+ cells in week 6 ATOs initiated with human cord blood (CB), adult bone marrow (BM), G-CSF mobilized peripheral blood (MPB), or non-mobilized peripheral blood (PB) HSPCs. (b) T cell progenitor subsets in ATOs from different HSPC sources, gated on CD34+ cells as shown in (a). (c) CD34+ progenitors and (d) CD34+ progenitor subsets in week 6 ATOs initiated with hematopoietic stem cell (HSC)-enriched (Lin-CD34+CD38-) fractions from the tissue sources shown. (e, f) Early onset of T cell commitment from LMPP and CD24- CLP in 3 week ATOs revealed by (e) early appearance of DP and (f) T cell committed CD34+CD7+ progenitors. Data are representative of two independent experiments.
Supplementary Figure 8 TCR diversity and functional validation of ATO-derived CD4+ T cells.
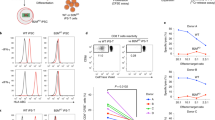
(a) Similar to human thymocytes, RAG1 and RAG2 are expressed in ATO-derived CD3+CD4+CD8+ (DP) but not mature CD3+CD8SP T cells. Quantitative RT-PCR for RAG1 and RAG2 are shown relative to expression of B2M in FACS sorted ATO-derived versus postnatal thymus T cell populations. Mean and SD of triplicate reactions is shown. (b) Generation of TCR diversity in CD3+TCRαβ+CD4SP T cells isolated from week 7 ATOs (n=5) or human thymi (n=4), as shown by flow cytometric analysis of the frequency of TCR Vβ family expression. (c) Cytokine production by week 12 ATO-derived CD4SP T cells treated with PMA/ionomycin for 6h. Data are representative of two independent experiments. (d) Proliferation (CTV dilution) and activation (upregulation of CD25) of cord blood (CB) and ATO-derived (week 12) CD4SP T cells after 5 days in response to anti-CD3/CD28 and IL-2. Data are representative of two individual experiments. (e) Post-ATO expansion of ATO-derived CD4SP T cells relative to starting cell number in response to anti-CD3/CD28 and IL-2 after 7 and 14 days. Mean and SD of technical triplicates are shown.
Supplementary Figure 9 Differentiation and allelic exclusion of TCR-engineered T cells in ATOs.
(a) ATO-derived TCR-engineered T cells retain a conventional T cell phenotype despite expansion and re-stimulation. CB HSPCs were transduced with an HLA-A*0201/NY-ESO-1157-165 specific TCR and then cultured in ATOs. After 6 weeks, CD8SP T cells were isolated from ATOs and activated with anti-CD3/28 beads + IL-2, expanded in IL-2, and re-stimulated with anti-CD3/28 beads on day 14. Preserved surface co-expression of CD8α and CD8β was confirmed by flow cytometry. Data are representative of two independent experiments. (b) Flow cytometric Vβ analysis of CD3+TCRαβ+tetramer+CD8SP T cells from TCR-transduced CB ATOs. Data are representative of 5 independent experiments (shown in graphical form in Fig. 5g). (c) Generation of TCR-engineered T cells from TCR-transduced CB HSPCs in ATOs using an HLA-A*02:01/MART126-35 specific TCR. Differentiation at week 6 is shown (gated on total CD14-CD56- ATO cells, with sequential gates shown above each plot). Data are representative of two independent experiments. (d) Antigen-specific priming of MART1-specific and NY-ESO-1-specific ATO-derived TCR-engineered T cells by artificial antigen presenting cells (aAPCs) that express CD80 and a HLA-A*02:01 single chain trimer presenting either MART126-35 or NY-ESO1156-165 peptide. CD107a membrane mobilization and intracellular IFNγ staining at 6h is shown. (e) In vitro cytotoxicity of ATO-derived TCR-engineered T cells against antigen-positive tumor cells. Frequencies of early (annexin V+ DAPI-) or late (annexin V+ DAPI+) apoptotic tumor cells was determined by flow cytometry at 9h (data are summarized in Fig 6h). (f) Retained antigen specificity following prolonged post-ATO activation/expansion of T cells. CD8SP T cells isolated from TCR-transduced ATOs were expanded for 14 days with anti-CD3/28 and IL-2, and cytotoxicity assays performed as described in panel (f) and Fig. 6h. Assays using TCR-transduced peripheral blood CD8+ donor T cells expanded for 14 days under the same conditions are shown for comparison.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 2366 kb)
Supplementary Protocol
DETAILED ARTIFICIAL THYMIC ORGANOID (ATO) METHOD (PDF 3240 kb)
Rights and permissions
About this article
Cite this article
Seet, C., He, C., Bethune, M. et al. Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nat Methods 14, 521–530 (2017). https://doi.org/10.1038/nmeth.4237
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4237
This article is cited by
-
Generation of complex bone marrow organoids from human induced pluripotent stem cells
Nature Methods (2024)
-
Parental Engagement in Identifying Information Needs After Newborn Screening for Families of Infants with Suspected Athymia
Journal of Clinical Immunology (2024)
-
Generating hematopoietic cells from human pluripotent stem cells: approaches, progress and challenges
Cell Regeneration (2023)
-
Base editing: a novel cure for severe combined immunodeficiency
Signal Transduction and Targeted Therapy (2023)
-
Bioengineering translational models of lymphoid tissues
Nature Reviews Bioengineering (2023)