Abstract
To expand the range of experiments that are accessible with optogenetics, we developed a photocleavable protein (PhoCl) that spontaneously dissociates into two fragments after violet-light-induced cleavage of a specific bond in the protein backbone. We demonstrated that PhoCl can be used to engineer light-activatable Cre recombinase, Gal4 transcription factor, and a viral protease that in turn was used to activate opening of the large-pore ion channel Pannexin-1.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tischer, D. & Weiner, O.D. Nat. Rev. Mol. Cell Biol. 15, 551–558 (2014).
Deisseroth, K. Nat. Neurosci. 18, 1213–1225 (2015).
Schröder-Lang, S. et al. Nat. Methods 4, 39–42 (2007).
Wu, Y.I. et al. Nature 461, 104–108 (2009).
Strickland, D. et al. Nat. Methods 9, 379–384 (2012).
Zhou, X.X., Chung, H.K., Lam, A.J. & Lin, M.Z. Science 338, 810–814 (2012).
Mizuno, H. et al. Mol. Cell 12, 1051–1058 (2003).
McEvoy, A.L. et al. PLoS One 7, e51314 (2012).
Kent, K.P., Oltrogge, L.M. & Boxer, S.G. J. Am. Chem. Soc. 131, 15988–15989 (2009).
Hoi, H. et al. J. Mol. Biol. 401, 776–791 (2010).
Shimizu-Sato, S., Huq, E., Tepperman, J.M. & Quail, P.H. Nat. Biotechnol. 20, 1041–1044 (2002).
Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. Nature 335, 563–564 (1988).
Sunaguchi, M., Nishi, M., Mizobe, T. & Kawata, M. Brain Res. 984, 21–32 (2003).
Picard, D. Methods Enzymol. 327, 385–401 (2000).
Scherrer, L.C. et al. Biochemistry 32, 5381–5386 (1993).
Matsuda, T. & Cepko, C.L. Proc. Natl. Acad. Sci. USA 104, 1027–1032 (2007).
Dempsey, W.P. et al. Nat. Methods 12, 645–648 (2015).
Kügler, J. et al. J. Biol. Chem. 287, 39224–39232 (2012).
Sandilos, J.K. et al. J. Biol. Chem. 287, 11303–11311 (2012).
Penuela, S., Gehi, R. & Laird, D.W. Biochim. Biophys. Acta 1828, 15–22 (2013).
Lohman, A.W. & Isakson, B.E. FEBS Lett. 588, 1379–1388 (2014).
Chekeni, F.B. et al. Nature 467, 863–867 (2010).
Weilinger, N.L. et al. Nat. Neurosci. 19, 432–442 (2016).
Wang, H. et al. Nat. Methods 13, 755–758 (2016).
Cheng, Z. & Campbell, R.E. ChemBioChem 7, 1147–1150 (2006).
Henderson, B.R. & Eleftheriou, A. Exp. Cell Res. 256, 213–224 (2000).
Kalderon, D., Roberts, B.L., Richardson, W.D. & Smith, A.E. Cell 39, 499–509 (1984).
Hathaway, N.A. et al. Cell 149, 1447–1460 (2012).
Potter, C.J., Tasic, B., Russler, E.V., Liang, L. & Luo, L. Cell 141, 536–548 (2010).
Campeau, E. et al. PLoS One 4, e6529 (2009).
Budhu, A. et al. Carcinogenesis 28, 1552–1560 (2007).
Hwang, D.R., Lai, H.Y., Chang, M.L., Hsu, J.T. & Yeh, C.T. J. Virol. Methods 129, 170–177 (2005).
Taremi, S.S. et al. Protein Sci. 7, 2143–2149 (1998).
Kim, J.H. et al. PLoS One 6, e18556 (2011).
Urbani, A. et al. J. Biol. Chem. 272, 9204–9209 (1997).
Roberts, P.J. et al. J. Biol. Chem. 283, 25150–25163 (2008).
Edelstein, A.D. et al. J. Biol. Methods 1, e10 (2014).
Shao, Q. et al. J. Biol. Chem. 291, 12432–12443 (2016).
Mohr, M.A., Argast, P. & Pantazis, P. Nat. Protoc. 11, 2419–2431 (2016).
Acknowledgements
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN 288338-2010 to R.E.C. and 435762-2013 to R.J.T.), the Canadian Institutes for Health Research (CIHR) (MOP-123514 to R.E.C. and 136812 to R.J.T.), and a Brain Canada Platform Support Grant. Additional support came from Alberta Innovates Technology Futures (AITF) (scholarship to W.Z.), the University of Alberta (scholarship to M.D.W.), the Alberta Ministry of Advanced Education (scholarship to Y.Z.), the Alberta Glycomics Centre (to J.S.K.), the Swiss National Science Foundation (SNF) (31003A 144048 to P.P.), and the European Union Seventh Framework Programme (Marie Curie Career Integration Grant (CIG) 334552 to P.P.). A.W.L. was supported by fellowships from CIHR and Alberta Innovates Health Solutions (AIHS). We thank G. Lambkin, K. Ballanyi, and Y. Ding for technical assistance, and S. Penuela (Western University), C. Cepko (Harvard Medical School), J. Crabtree (Stanford University), L. Luo (Stanford University), E. Campeau (University of Massachusetts), K. Deisseroth (Stanford University), X. Wang (National Cancer Institute), and D. Trono (École Polytechnique Fédérale de Lausanne) for providing reagents.
Author information
Authors and Affiliations
Contributions
W.Z., A.W.L., P.P., R.J.T., and R.E.C. conceived and designed experiments. W.Z. assembled all constructs and characterized protein in vitro. W.Z. and M.D.W. performed directed evolution. W.Z., X.L., and H.H. performed imaging of protein translocation. W.Z. and Y.Z. performed light-induced gene expression. A.W.L. characterized Opto-Panx1. S.Y., M.A.M., and P.P. performed primed conversion. E.N.K. and J.S.K. performed mass spectrometry analysis. All authors were involved in data analysis, and W.Z., A.W.L., R.J.T., and R.E.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
W.Z., H.H., and R.E.C. are inventors on a provisional patent application that describes PhoCl.
Integrated supplementary information
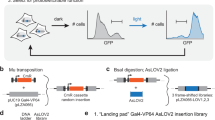
Supplementary Figure 1 Strategy for design and screening of PhoCl variants.
(a) Schematic of circular permutation libraries. The gene encoding mMaple1 was circularly permuted at six surface exposed positions either N- or C-terminal to the central helix. For each permutation, libraries were constructed by randomizing both the N-terminal residue (X) and the C-terminal residue (Y) to all 20 amino acid possibilities, to give a total of 400 possible variants. For circularly permuted variants the residues are numbered as in mMaple (Supplementary Fig. 3) and the C-terminal residue (Y) is the residue that preceded the N-terminal residue (X) in mMaple. (b) Schematic of colony-based screening of libraries of PhoCl variants. PhoCl libraries were screened in colonies of E. coli (each expressing a library variant). Petri dishes containing hundreds of colonies were imaged (green and red fluorescence), illuminated in a 405 nm LED chamber, imaged immediately after being removed from the chamber, and then imaged again after 10 min at room temperature. Colonies that exhibited bright red fluorescence immediately after photoconversion, and then lost this fluorescence within 5 – 10 min at room temperature, were picked for further investigation. (c) Screening of libraries constructed as in a, using the method represented in b, led to the identification of PhoCl0.1 with N-terminal residue D79V and C-terminal residue E78R. As shown in this 2D topology representation of PhoCl (based on the structure of mTFP1 (PDB ID 2HQK)2), the permutation site is on the C-terminal side of the central helix. The yellow dashed line represents the GGSGG linker connecting the original N- and C-termini. Residues 53-56, where permutation led to photoconvertible proteins that did not spontaneously dissociate, are indicated in magenta.
1. McEvoy, A.L. et al. PLoS ONE 7, e51314 (2012).
2. Ai, H.W., Henderson, J.N., Remington, S.J. & Campbell, R.E. Biochem. J. 400, 531-540 (2006).
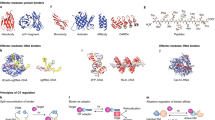
Supplementary Figure 2 Representation of PhoCl structure and lineage of improved variants.
(a) Structural representation of PhoCl dissociation modeled on the structure of mTFP1 (PDB ID 2HQK)1. As photocleavage occurs on the N-terminal side of the chromophore, and the permutation site is on the C-terminal side of the chromophore, the small peptide that dissociates from the larger fragment contains the chromophore. The fluorescence of the chromophore is quenched outside of the β-barrel. The small and large fragments are colored magenta and green, respectively. (b) Lineage of improved PhoCl variants discovered during iterative cycles of library screening using the approach shown in Supplementary Fig. 1b. From PhoCl0.1 on, libraries were generated by error-prone PCR. The I15N and F178Y mutations provided the most substantial increases in the rate of peptide dissociation. (c) PhoCl mutations (orange) modeled on the structure of mTFP1 (PDB ID 2HQK). Small and large fragments resulting from photocleavage are colored magenta and green, respectively. M6dT is not visible in the mTFP1 crystal structure and is therefore not represented here.
1. Ai, H.W., Henderson, J.N., Remington, S.J. & Campbell, R.E. Biochem. J. 400, 531-540 (2006).
Supplementary Figure 3 Sequence alignment of mMaple and PhoCl variants.
Mutations relative to mMaple1 are represented as white text on a black background. The chromophore-forming residues are represented as black text on a green background. The circular permutation (cp) linker sequence is enclosed in a box. The site of photocleavage is indicated with an arrow.
1. McEvoy, A.L. et al. PLoS ONE 7, e51314 (2012).
Supplementary Figure 4 Absorbance and fluorescence spectra of mMaple and PhoCl.
(a) Normalized fluorescence excitation (dashed line) and emission (solid line) spectra for the initial green state of PhoCl. (b) Normalized absorbance (black to gray lines) and fluorescence emission (magenta line) spectra for PhoCl post photoconversion. Fluorescence emission was acquired less than 30 s after a 5 min photoconversion in the 405 nm LED chamber. The “0 min” absorbance spectrum was acquired less than 10 s after photoconversion. Subsequent spectra were acquired 5, 10, 15, and 20 min later. Most of the protein had dissociated prior to the first measurement. (c) Normalized absorbance spectra for mMaple1 before (green) and after (magenta) photoconversion. (d) Normalized fluorescence excitation (dashed) and emission (solid) for the initial (green lines) and photoconverted (magenta lines) states of mMaple.
1. McEvoy, A.L. et al. PLoS ONE 7, e51314 (2012).
Supplementary Figure 5 Demonstration of PhoCl photocleavage by mass spectrometry and western blotting.
(a) Schematic representation of illumination and time-lapse electrospray mass spectrometry (ESI-MS) data acquisition. PhoCl was photoconverted in the 405 nm LED chamber (0.15 mW/mm2) with a 2 min illumination starting at t = 0 s, and a 6 min illumination starting at t = 2015 s. ESI mass spectra were acquired every 1 s for 30 min starting at t = 215 s, and for 10 min starting at t = 2500 s. (b) ESI mass spectra of PhoCl acquired at t = 0 s (before photoconversion), 215 s, 2015 s and 3100 s. Percent cleavage was calculated as EB/(EB + PhoCl) × 100, where EB and PhoCl are the sum of the peak heights for various corresponding ionization states. (c) Percent cleavage versus time, from t = 215 s to t = 2015 s. (d) Quantitative western blot of PhoCl-mCherry-myc photocleavage following 405 nm illumination (2, 4, 6, 8 min at 0.15 mW/mm2). Membranes were probed with an anti-myc primary antibody. Violet light induced a band shift from ~60 kD (full length PhoCl-mCherry-myc) to ~30 kD (mCherry-myc) indicating the cleavage of PhoCl. Values are means ± s.d. (n = 3).
Supplementary Figure 6 Optogenetic manipulation of protein localization with PhoCl.
(a) Representative fluorescence image of HeLa cells expressing NES-PhoCl-mCherry before photoconversion. Regions of interest used to quantify fluorescence intensity changes are indicated as follows: nucleus (region outlined in red), cytoplasm (region outlined in white minus region outlined in yellow), and the whole cell (region outlined in white). Scale bar, 20 μm. (b) Normalized and averaged whole cell red fluorescence intensity versus time for the cells in a. Light pulses are 5 s in duration, every 15 s for 6 min. Values are means ± s.d. (n = 3). (c) Photoconversion of PhoCl (quantified by the loss of whole cell green fluorescence) and red fluorescence intensity localization ratio (cytoplasm to nucleus) versus time, for the cells in a. Values are means ± s.d. (n = 3). (d) Representative fluorescence image of HeLa cells expressing NLS-PhoCl-mCherry before photoconversion. Regions of interest are defined as in a. Scale bar, 20 μm. (e) Normalized and averaged whole cell red fluorescence intensity versus time for the cells in d. Illumination conditions are as described in b. Values are means ± s.d. (n = 3). (f) Photoconversion of PhoCl (quantified by the loss of whole cell green fluorescence) and red fluorescence intensity localization ratio (nucleus to cytoplasm) versus time, for the cells in d. Values are means ± s.d. (n = 3).
Supplementary Figure 7 Optogenetic manipulation of protein localization in a single cell.
(a) Time-lapse images of HeLa cells expressing NLS-PhoCl-mCherry before and after 15 s illumination of cell 1 (arrow) with a 405 nm laser (0.12 mW). Saturated pixels in the mCherry channel with 3-fold increased contrast are shown in white. Scale bar, 20 μm. (b) Normalized whole cell green fluorescence intensity versus time for the cells in a. (c) Red fluorescence localization ratio (nucleus intensity to cytoplasm intensity) versus time, for the cells in a.
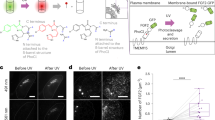
Supplementary Figure 8 Optogenetic redistribution of the N- and C-terminal portions of NES-GFP-PhoCl-mCherry-NLS after photocleavage and in situ demonstration of PhoCl photocleavage with ddFPs.
(a) GFP with a NES and mCherry with an NLS were genetically fused to the N- and C-termini, respectively, of PhoCl. Upon photocleavage the two FPs are released from each other and the GFP is now excluded from the nucleus, while the red FP is sequestered in the nucleus. (b) Ratiometric images (red/green) of HeLa cells expressing the construct described in a before and after illumination with ~400 nm light. Before illumination with 400 nm light, the protein is largely excluded from the nucleus (inset of leftmost image) but the ratio of red to green fluorescence is generally consistent throughout the nucleus and cytoplasm. After photocleavage the ratio of red to green fluorescence decreases in the cytoplasm and increases in the nucleus. Scale bar, 15 μm. (c) Demonstration of PhoCl photocleavage in live cells using dimerization dependent FP (ddFP) technology1,2. “RA” is a dimly fluorescent red FP that becomes brighter upon heterodimerization with its non-fluorescent homolog “RB”. As with NES-GFP-PhoCl-mCherry-NLS in b, the NES-PhoCl-RB-NLS construct is initially excluded from the nucleus. Upon photocleavage, the RB-NLS portion is separated from the NES and translocates to the nucleus where it heterodimerizes with RA, causing an increase in red fluorescence. (d) Green and red fluorescence images of cells expressing the constructs described in c before and after illumination with ~400 nm light. Within 20 min of illumination, the red fluorescence in the nucleus is observed to increase due to translocation and interaction of RA with RB. Regions of interest used to quantify fluorescence intensity changes are indicated as follows: nucleus (outlined in red) and cytoplasm (outlined in white). Scale bar, 20 μm. (e) Nucleus-to- cytoplasm intensity ratio for cells in d. Values are means ± s.d. (n = 4).
1. Alford, S.C., Abdelfattah, A.S., Ding, Y. & Campbell, R.E. Chem. Biol. 19, 353-360 (2012).
2. Alford, S.C., Ding, Y., Simmen, T. & Campbell, R.E. ACS Synth. Biol. 1, 569-575 (2012).
Supplementary Figure 9 Optogenetic release of membrane-tethered Gal4 transcription factor to induce gene expression.
(a) Schematic representation of activation mechanism. PhoCl was genetically inserted between the Gal4-VP16 transcription factor1, with or without an NLS, and the plasma-membrane associated β-2 adrenergic receptor (β2AR)2. The resulting plasmid (β2AR-PhoCl-Gal4-VP16), together with a reporter plasmid encoding mCherry-NLS under control of a repeated upstream activator sequence (UAS)3, was used to cotransfect mammalian cells. Photocleavage would allow Gal4-VP16 to translocate to the nucleus, bind to the UAS promoter, and activate gene expression. (b) Green (PhoCl) and red (mCherry) fluorescence images of a HeLa cell that has been transfected with the genes encoding β2AR-PhoCl-Gal4-VP16 and pUAS-mCherry-NLS, and has not been illuminated with 400 nm light. The strong mCherry-NLS expression in the absence of illumination reveals a high level of transcription even though β2AR-PhoCl-Gal4-VP16 does appear to be membrane localized and excluded from the nucleus. Scale bar, 20 μm. (c) Red fluorescence images of HeLa cells expressing β2AR-mCherry (left image) and both β2AR-Gal4-VP16 and pUAS-mCherry-NLS (right image), and not illuminated with 400 nm light. These control experiments confirm the membrane localization of β2AR and that mCherry-NLS expression occurs even with a membrane tethered Gal4-VP16 construct that does not contain PhoCl. Scale bars, 20 μm.
1. Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. Nature 335, 563-564 (1988).
2. Sunaguchi, M., Nishi, M., Mizobe, T. & Kawata, M. Brain Res. 984, 21-32 (2003).
3. Southall, T.D., Elliott, D.A. & Brand, A.H. CSH Protoc. 2008, pdb.top49 (2008).
Supplementary Figure 10 Optogenetic manipulation of gene expression with PhoCl.
(a) Quantification of light-activated gene expression with SR-PhoCl-Gal4-VP16-PhoCl-SR and an mCherry reporter. Cells were transiently transfected with the constructs indicated, illuminated as in a, and then analyzed 24 hours after illumination. Values are means ± s.d. (n = 3). (b) Quantification of light-activated gene expression with SR-PhoCl-Gal4-VP16-PhoCl-SR and a NanoLuc reporter. Cells were transiently transfected with the constructs indicated and analyzed 12 hours after illumination (2 min of 405 nm at ~0.15 mW/mm2, repeated at 24, 25, and 26 h post transfection). Values are means ± s.d. (n = 3). (c) Quantification of light-activated recombination with SR-PhoCl-Cre-PhoCl-SR and a floxed mCherry reporter. Cells were transiently transfected with the constructs indicated, illuminated as in a, and then analyzed 24 hours after illumination. mCherry fluorescence increased 46 ± 8-fold with illumination. Values are means ± s.d. (n = 3). (d) Flow cytometry analysis to assess optogenetic activation of Cre recombinase. Representative histograms show distribution of mCherry fluorescence intensity in a HEK 293 stable cell line (containing a floxed inverted mCherry reporter gene) that has been transiently transfected with the constructs indicated, with and without illumination (as in a). The range of intensities considered to be mCherry-positive cells is indicated with a blue line. Values are means ± s.d. (n = 3). Histograms with the Y-axis (counts) zoomed in are presented in the bottom row.
Supplementary Figure 11 Primed conversion in vitro and in live cells.
(a) Primed conversion1,2 of PhoCl-MBP embedded in a polyacrylamide gel. Green and red PhoCl fluorescence signal after attempted 458 nm photoconversion (left), 730 nm photoconversion (center), and primed conversion using 458 nm and 730 nm (right). Merged channels are shown on the bottom row. Laser powers were both set to 100%. Scale bar, 50 μm. (b) Change in green (green bars) and red (magenta bars) fluorescence intensity of PhoCl-MBP upon primed conversion using a 458 nm priming beam at varying laser powers and constant 100% 730 nm laser. Values are means ± s.d. (n = 3). (c) Fluorescence microscopy images of HEK 293T cells co-transfected with SR-PhoCl-Gal4-VP16-PhoCl-SR and pUAS-mCherry-NLS. Before illumination (left), after 458 nm laser only illumination on upper cell (center), and primed conversion on the lower cell (right). Scale bar, 10 μm. (d) Quantification of red nuclei over total number of green cells treated as in c. Cells were either subjected to primed conversion using 458 nm plus 730 nm beams (red bar), kept in dark (grey bar), illuminated with only 458 nm (white bar), or illuminated with 405 nm (blue bar). Values are means ± s.d. (n = 3).
1. Mohr, M.A. & Pantazis, P. Methods Cell Biol. 133, 125-138 (2016).
2. Mohr, M.A., Argast, P. & Pantazis, P. Nat. Protoc. 11, 2419-2431 (2016).
Supplementary Figure 12 Identification of an optimal inhibitory peptide (I) for I-PhoCl-HCVp.
(a) Schematic of I-PhoCl-HCVp and I-p2A-HCVp constructs. (b) Structural representation of I-PhoCl-HCVp. HCVp (cyan) in represented in complex with the inhibitory peptide CP5-46A-4D5E (red) (PDB ID 4A1V)1, and PhoCl (green) is represented using the structure of mTFP1 (PDB ID 2HQK)2. (c) Structural representation of I-p2A-HCVp, where p2A is a self-cleaving peptide3. (d) Amino acid sequences of five peptide inhibitors, with Ki values ranging from 0.5-100 nM1. (e) Representative fluorescence microscopy images (mCherry channel) of HEK 293 cells co-expressing I-p2A-HCVp plus mCherry-4A/4B-CAAX (top row) or I-PhoCl-HCVp plus mCherry-4A/4B-CAAX (bottom row). None of the cells are illuminated with 400 nm light to induce photocleavage. In each column, I is the same for both I-p2A-HCVp and I-PhoCl-HCVp, and corresponds to one of the sequences provided in c. Scale bars, 10 μm.
1. Kügler, J. et al. J. Biol. Chem. 287, 39224-39232 (2012).
2. Ai, H.W., Henderson, J.N., Remington, S.J. & Campbell, R.E. Biochem. J. 400, 531-540 (2006).
3. Kim, J.H. et al. PLoS ONE 6, e18556 (2011).
Supplementary Figure 13 Engineering of HCVp-activatable Panx1 (Panx1HCV) and validation by dye uptake assay.
(a) Partial amino acid sequence showing the caspase-3 substrate sequence within Panx1WT (Ref. 1). This sequence was substituted with an HCVp substrate sequence to make Panx1HCV. (b) Representative western blot showing Opto-Panx1 C-tail cleavage upon violet light stimulation (405 nm, 0.15 mW/mm2). Panx1-EL denotes a primary antibody against the first extracellular domain of Panx1 (total Panx1). Panx1-CT denotes a primary antibody directed against an epitope distal to the HCVp site in the C-terminal domain of Panx1HCV (uncleaved Panx1). NT = non-transfected, Panx1HCV = Panx1HCV only, Opto-Panx1 = I-PhoCl-HCVp and Panx1HCV. (c) Quantification of band density ratio (uncleaved Panx1 to total Panx1) in b normalized to the ratio for cells expressing Panx1HCV only. Values are means ± s.d. (n = 3). (d) Expression of Opto-Panx1 (I-PhoCl-HCVp and Panx1HCV-mCherry) in N2a cells. Scale bar, 10 μm. (e) Quantification of dye in cells expressing Panx1HCV and either I-PhoCl-HCVp (Opto-Panx1) or I-mMaple-HCVp, when activated using a 380 nm LED (purple interval). Values are means ± s.d. (nOpto-Panx1 = 26, nI-mMaple-HCVp:Panx1HCV = 40), P20 min = 1.9 × 10-9, P25 min = 3.6 × 10-14, P30 min < 1 × 10-15 for Opto-Panx1 compared to I-mMaple-HCVp:Panx1HCV by two-way ANOVA (F (7, 448) = 25.9). (f) Time-lapse images of TO-PRO-3 dye uptake resulting from 3 min 380 nm light activation of Opto-Panx1 in N2a cells. Inset in leftmost PhoCl image shows Panx1HCV-mCherry expression. Scale bar, 10 μm.
1. Sandilos, J.K. et al. J. Biol. Chem. 287, 11303-11311 (2012).
Supplementary Figure 14 Whole-cell patch-clamp analysis of cells expressing Opto-Panx1 or the mMaple control construct.
(a) Representative current (I) versus voltage (V) curves for whole-cell patch clamp analysis of the negative control construct I-mMaple-HCVp:Panx1HCV in HEK 293T cells. (b, c) Representative current versus time data for whole-cell patch clamp analysis of HEK 293T cells expressing Panx1HCV and either I-mMaple-HCVp (b) or I-PhoCl-HCVp (Opto-Panx1) (c). Cells were illuminated with a 380 nm LED for 1 min (purple interval) and treated with carbenoxolone (CBX: 100 μm after 10 min (red interval). (d) Summary data of light-induced Panx1 currents at +80 mV. Values are means ± s.d. (nOpto-Panx1 = 8, nI-mMaple-HCVp:Panx1HCV = 8).
Supplementary Figure 15 Blebbing analysis of cells expressing Opto-Panx1 or the mMaple control construct.
(a) Representative time course images of N2a cells expressing Opto-Panx1 following 3 min of illumination with a 380 nm LED. Blebs are indicated with yellow arrowheads. Scale bar, 10 μm. (b) Summary of number of blebs per cell following activation of Opto-Panx1 or I-mMaple-HCVp:Panx1HCV in N2a cells. Values are means ± s.d. (nOpto-Panx1 = 26, nI-mMaple-HCVp:Panx1HCV = 40). P = 3.3 × 10-12 by unpaired Student’s t-test (t (64) = 8.6). (c) Average bleb area per cell for data in b. P = 1.6 × 10-11 by unpaired Student’s t-test (t (64) = 8.2).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Note and Supplementary Table 1 (PDF 3604 kb)
Source data
Rights and permissions
About this article
Cite this article
Zhang, W., Lohman, A., Zhuravlova, Y. et al. Optogenetic control with a photocleavable protein, PhoCl. Nat Methods 14, 391–394 (2017). https://doi.org/10.1038/nmeth.4222
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4222
This article is cited by
-
An optogenetic method for the controlled release of single molecules
Nature Methods (2024)
-
A single-component, light-assisted uncaging switch for endoproteolytic release
Nature Chemical Biology (2024)
-
Irreversible light-activated SpyLigation mediates split-protein assembly in 4D
Nature Protocols (2024)
-
Real-time tracking of bioluminescent influenza A virus infection in mice
Scientific Reports (2022)
-
Efficient spatially targeted gene editing using a near-infrared activatable protein-conjugated nanoparticle for brain applications
Nature Communications (2022)