Abstract
We introduce Cryogenic Optical Localization in 3D (COLD), a method to localize multiple fluorescent sites within a single small protein with Angstrom resolution. We demonstrate COLD by determining the conformational state of the cytosolic Per-ARNT-Sim domain from the histidine kinase CitA of Geobacillus thermodenitrificans and resolving the four biotin sites of streptavidin. COLD provides quantitative 3D information about small- to medium-sized biomolecules on the Angstrom scale and complements other techniques in structural biology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Betzig, E. Opt. Lett. 20, 237–239 (1995).
Weisenburger, S. & Sandoghdar, V. Contemp. Phys. 56, 123–143 (2015).
Zondervan, R., Kulzer, F., Kolchenko, M. & Orrit, M. J. Phys. Chem. A 108, 1657–1665 (2004).
Weisenburger, S., Jing, B., Renn, A. & Sandoghdar, V. In Proc. SPIE 8815, 88150D (SPIE, 2013).
Li, W., Stein, S.C., Gregor, I. & Enderlein, J. Opt. Express 23, 3770–3783 (2015).
Weisenburger, S. et al. Chemphyschem 15, 763–770 (2014).
Sevvana, M. et al. J. Mol. Biol. 377, 512–523 (2008).
Cheng, Y., Grigorieff, N., Penczek, P.A. & Walz, T. Cell 161, 438–449 (2015).
Dvornek, N.C., Sigworth, F.J. & Tagare, H.D. J. Struct. Biol. 190, 200–214 (2015).
Harauz, G. & van Heel, M. Optik (Stuttg.) 73, 146–156 (1986).
Rice, S.O. Bell Labs Tech. J. 24, 46–156 (1945).
Jung, J. et al. Microsc. Res. Tech. 76, 835–843 (2013).
Courtois, J., Courty, J. & Mertz, J.C. Phys. Rev. A 53, 1862–1878 (1996).
Hettich, C. et al. Science 298, 385–389 (2002).
Lippitz, M. et al. Phys. Rev. Lett. 92, 103001 (2004).
Taraska, J.W., Puljung, M.C., Olivier, N.B., Flynn, G.E. & Zagotta, W.N. Nat. Methods 6, 532–537 (2009).
Shevelev, G.Y. et al. J. Phys. Chem. B 119, 13641–13648 (2015).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Sheldrick, G.M. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 (2010).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Nat. Protoc. 3, 1171–1179 (2008).
Murshudov, G.N. et al. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Niu, T., Ding, A.A., Kreutz, R. & Lindpaintner, K. Genetics 169, 1021–1031 (2005).
Do, C.B. & Batzoglou, S. Nat. Biotechnol. 26, 897–899 (2008).
Kolmakov, K. et al. European J. Org. Chem. 2010, 3593–3610 (2010).
Pettersen, E.F. et al. J. Comput. Chem. 25, 1605–1612 (2004).
van Heel, M. & Schatz, M. J. Struct. Biol. 151, 250–262 (2005).
Acknowledgements
This work was supported by the Max Planck Society and the Alexander von Humboldt Foundation (Humboldt Professor, V.S.). We thank L. Wei for her contribution to the earlier stage of this work. We thank E. Eichhammer, S. Götzinger and T. Utikal for their contributions in the development of the cryostat extension. We also thank I. Schoen and M. Piliarik for fruitful discussions. The work on the PAS domain was supported by the DFG (to C.G., grant no. GR1211/17-1). S.B. thanks G. Sheldrick for advice on refinement using data from twinned crystals. We thank G. Unden (University of Mainz, Germany) for the GtCitA clone.
Author information
Authors and Affiliations
Contributions
V.S. conceived and supervised the project. S.W. built the experimental setup. S.W. and D.B. performed the optical measurements and analyzedthe data. B.S., K.G. and S.B. provided GtCitA PASc samples and solved the crystal structure. B.S. and C.G. recorded and analyzed NMR spectra. S.W. and V.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Control experiments on the fluorescence stability of single Atto647N-biotin complexes.
a, Exemplary signal trace (black line) in counts per second (cps) of a single Atto647N molecule attached to a single biotin molecule with linear combination fit (N = 1, red dots). b, Plot of the difference between the fit and the experimental trace, yielding RMS = 37 cps. The fit error is 2.9 times the shot noise. The integration time per point was chosen to be 2 seconds as an empirical compromise to minimize fluctuations due to fast blinking events (as in our other measurements in the manuscript). c, Histogram of the shot-noise-corrected fit error for many single Atto647n-biotin molecules.
Supplementary Figure 2 Fluorophore on-times and photon numbers.
On-time photoblinking statistics at liquid helium temperature for individual Atto647N molecules coupled to PASc (a) and coupled to streptavidin via biotin (b). Histograms of the detected number of photons per fluorophore for the PASc domain dimer (c) and the streptavidin molecule (d). The histograms show the detected number of photons per fluorophore on a protein complex. Since in the case of streptavidin there are possible intensity combinations, each molecule is less often observed alone. This reduces the average total number of detected photons per fluorophore during the same observation time.
Supplementary Figure 3 Intensity level allocation.
a, Visualization of the uncertainty in determining the position of each intensity level for an example streptavidin recording. Normal distributions (various colors) are plotted at the determined positions with their widths encoding the experimentally observed fit error. The variables of the fitting procedure (intensity levels of the individual fluorophores I1..I4) are marked as well as some linear combination levels. b, Probability of the correct allocation of a measured intensity to the estimated intensity level as a function of the number of fluorophores.
Supplementary Figure 4 Complete data sets from the PASc domain dimer measurements.
Data for localization precisions better than 6.5 Å. Black crosses indicate the measured positions while the precision is encoded as the width of the 2D Gaussian probability distributions. Plots are 5 x 5 nm.
Supplementary Figure 5 Crystal structure of GtPASc.
a, Crystal structure of GtPASc reveals a dimeric arrangement of monomers (green and grey) in which the N-terminal helices are swapped. b, The view from (a) is rotated by 90° for better visualization of N-terminal helix packing.
Supplementary Figure 6 Fourier shell correlation of two half data sets.
a, For the PASc domain dimer. The half-bit criterion yields a resolution of 1.9 Å. b, For the streptavidin complex. The half-bit criterion yields a resolution of 5.0 Å. The FSC was computed using Free FSC (IMAGIC).
Supplementary Figure 7 NMR on labeled PASc.
a, Overlay of 1H,15N-HSQC-spectra of 1 mM GtCitA PASc (blue) with 25 μM Atto647N-labeled PASc N308C (red). Peaks for unfolded PASc after dye labeling were not observable. Peaks corresponding to the β-scaffold are partially broadened out due to anisotropic effects from the dye molecules. b, Proton chemical shift difference of identified residues in Atto647N-labeled PASc compared to unlabeled PASc reference shifts. Only minor chemical shift changes are visible; an observed peak doubling for some residues (red asterisks) is likely caused by the achieved dye/protein ratio of 0.65, resulting in different molecular species in the NMR sample to be detected. c, Proton chemical shift differences of more than 0.03 ppm (red) mapped on the PASc crystal structure. Dye attachment site is shown in green. d, Chemical structure of the Atto647N dye molecule attached to the C-termini.
Supplementary Figure 8 Complete data sets from the streptavidin measurements.
Data for localization precisions better than 13 Å. Black crosses indicate the measured positions while the precision is encoded as the width of the 2D Gaussian probability distributions. Plots are 5 x 5 nm.
Supplementary Figure 9 Protein orientation for streptavidin.
Data sets form Fig. 3 (a-d) and additional data sets (e–h). Shown are the measurements, least squares fit results to the projection of a model volume, the fit residuals of the fit to the projection of a model volume, and least-squares fit results to projections of the reconstructed volume. See the Methods section for details.
Supplementary Figure 10 Schematic of the cryogenic and optical setup.
Schematic of the beam path and optical components. BP - band pass filter; DM - dichroic mirror; QD - quad diode; λ/2 - half wave plate; λ/4 - quarter wave plate; WF lens - wide-field lens; LP - long pass filter; CCD - CCD camera. See the Methods section for a detailed description of the experimental setup.
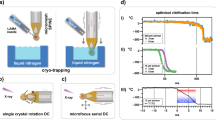
Supplementary Figure 11 Basics of COLD microscopy.
a, A commercial flow cryostat (lower part of the drawing) extended by a vacuum chamber (upper part; the walls are not shown for clarity) houses a microscope objective with a numerical aperture of 0.9. The sample is placed on a cold finger under the objective. See Methods for more details. b, Photograph of the cryostat with the vacuum chamber extension. c, Example of a wide-field fluorescence image recorded at T = 4.3 K with an integration time of 2 s. d, Zoom of one diffraction-limited spot.
Supplementary Figure 12 Flow diagram describing the single-molecule data analysis.
See the Online Methods for a detailed description of the analysis.
Supplementary Figure 13 Systematic localization error.
a, Geometry for the simulations (see Supplementary Note 1). The molecules have a polar angle θ and a distance to the interface between the two media, z. Detection through the low-index medium. b, Simulation of the error in the localization accuracy as a function of z and θ, assuming NA = 0.9.
Supplementary Figure 14 Drift correction.
a,b, Example trace for the estimated localizations of the individual fluorophores (various colors). Weighted mean position for the molecule of interest in black. Estimated positions for all localizations from four fluorophores coupled to a molecule before (c) and after drift correction (d). Crosses mark the final positions of the four fluorophores. The sigma of the spread of individual localizations is on the order of 5-10 nm in accordance with our localization precision per frame, yielding the final localization precisions shown in Figs. 2c and 3c.
Supplementary Figure 15 Euler angle convention.
Rotations about the angles α, β and γ transform the coordinate system xyz into x’y’z’.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1 and 2, and Supplementary Note.
Supplementary Protocol
COLD measurements.
Increasing the quality of the 3D reconstruction with increasing the quality of the input data.
Demonstration of the influence of the localization precision of the projection data on the 3D reconstruction. We gradually increase the requirement on the data quality by changing the filtering conditions from using only data sets with 20 Å < σLoc < 30 Å to using the best data sets with σLoc < 13 Å while keeping the number of input images similar. The movie shows an overlay of the reconstructed volumes (red) with the crystal structure. Isosurfaces are plotted at an isovalue of 0.68.
Raw data recording.
Example of a wide-field fluorescence image stack recorded at T = 4.3 K with an integration time of 2 s. The movie shows the first hour raw data from a 2-hour recording. A black frame every 15 min indicates when automated refocusing is performed. Brightness has been adjusted for clarity.
Source data
Rights and permissions
About this article
Cite this article
Weisenburger, S., Boening, D., Schomburg, B. et al. Cryogenic optical localization provides 3D protein structure data with Angstrom resolution. Nat Methods 14, 141–144 (2017). https://doi.org/10.1038/nmeth.4141
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4141
This article is cited by
-
Three-dimensional magnetic resonance tomography with sub-10 nanometer resolution
npj Quantum Information (2024)
-
MINSTED nanoscopy enters the Ångström localization range
Nature Biotechnology (2023)
-
Superresolution concentration measurement realized by sub-shot-noise absorption spectroscopy
Nature Communications (2022)
-
Single-molecule localization microscopy
Nature Reviews Methods Primers (2021)
-
3D particle averaging and detection of macromolecular symmetry in localization microscopy
Nature Communications (2021)