Abstract
We describe a red-shifted fluorescence resonance energy transfer (FRET) pair optimized for dual-color fluorescence lifetime imaging (FLIM). This pair utilizes a newly developed FRET donor, monomeric cyan-excitable red fluorescent protein (mCyRFP1), which has a large Stokes shift and a monoexponential fluorescence lifetime decay. When used together with EGFP-based biosensors, the new pair enables simultaneous imaging of the activities of two signaling molecules in single dendritic spines undergoing structural plasticity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
NCBI Reference Sequence
Referenced accessions
NCBI Reference Sequence
References
Jares-Erijman, E.A. & Jovin, T.M. Nat. Biotechnol. 21, 1387–1395 (2003).
Zhai, S., Ark, E.D., Parra-Bueno, P. & Yasuda, R. Science 342, 1107–1111 (2013).
Murakoshi, H., Wang, H. & Yasuda, R. Nature 472, 100–104 (2011).
Lee, S.J., Escobedo-Lozoya, Y., Szatmari, E.M. & Yasuda, R. Nature 458, 299–304 (2009).
Nishiyama, J. & Yasuda, R. Neuron 87, 63–75 (2015).
Chu, J. et al. Nat. Biotechnol. 34, 760–767 (2016).
Shaner, N.C., Steinbach, P.A. & Tsien, R.Y. Nat. Methods 2, 905–909 (2005).
Costantini, L.M., Fossati, M., Francolini, M. & Snapp, E.L. Traffic 13, 643–649 (2012).
Cranfill, P.J. et al. Nat. Methods 13, 557–562 (2016).
Mainen, Z.F. et al. Methods 18, 231–239 (1999).
Chen, T.W. et al. Nature 499, 295–300 (2013).
Katayama, H., Yamamoto, A., Mizushima, N., Yoshimori, T. & Miyawaki, A. Cell Struct. Funct. 33, 1–12 (2008).
Murakoshi, H., Lee, S.J. & Yasuda, R. Brain Cell Biol. 36, 31–42 (2008).
Chu, J. et al. Nat. Methods 11, 572–578 (2014).
Bajar, T.B. et al. Nat. Methods http://dx.doi.org/10.1038/nmeth.4045.
Matsuzaki, M., Honkura, N., Ellis-Davies, G.C. & Kasai, H. Nature 429, 761–766 (2004).
Yasuda, R. et al. Sci. STKE 2004, pl5 (2004).
Murakoshi, H., Shibata, A.C., Nakahata, Y. & Nabekura, J. Sci. Rep. 5, 15334 (2015).
Chalfie, M. & Kain, S.R. (eds.) Green Fluorescent Protein: Properties, Applications and Protocols (John Wiley & Sons, Inc., 1998).
Wilkins, M.R. et al. Methods Mol. Biol. 112, 531–552 (1999).
Lam, A.J. et al. Nat. Methods 9, 1005–1012 (2012).
Stoppini, L., Buchs, P.A. & Muller, D. J. Neurosci. Methods 37, 173–182 (1991).
O'Brien, J.A. & Lummis, S.C. Nat. Protoc. 1, 977–981 (2006).
Saito, T. Nat. Protoc. 1, 1552–1558 (2006).
Holtmaat, A. et al. Nat. Protoc. 4, 1128–1144 (2009).
Pologruto, T.A., Sabatini, B.L. & Svoboda, K. Biomed. Eng. Online 2, 13 (2003).
Acknowledgements
We would like to thank D. Kloetzer for lab management, L. Yan for help with FLIM microscopy setup and technical assistance, J. Richards for preparation of slice cultures, L. Colgan for comments on the manuscript, W. Jiang and E. Mellins for assistance with size-exclusion chromatography, J. Chu for assistance with molecular biology and protein characterization, and members of the Lin laboratory for technical assistance. This work was supported by grants from Human Frontiers Science Program (long-term fellowship to T.L.); NSF Graduate Fellowship (B.B.K.); a Siebel Scholar Award (A.J.L.); National Institute of Health grants R01MH080047 and 1DP1NS096787 to R.Y. and 1U01NS090600 and P50GM107615 to M.Z.L.; a Burroughs Wellcome Foundation Career Award for Medical Scientists (M.Z.L.); a Rita Allen Foundation Scholar Award (M.Z.L.); and the Max Planck Florida Institute for Neuroscience.
Author information
Authors and Affiliations
Contributions
T.L. performed FLIM characterization, FLIM imaging in slices, and in vivo imaging. J.C. initially engineered and characterized CyRFP1; B.B.K. engineered mCyRFP1 and characterized new variants. A.J.L. engineered mMaroon1. M.Z.L. and R.Y. provided supervision. T.L., M.Z.L., and R.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
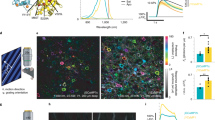
Supplementary Figure 1 Characterization of CyRFP1 and mCyRFP1.
(a) Size-exclusion chromatography of EGFP, tdTomato, mCherry, CyOFP1, CyRFP1, and mCyRFP1. EGFP and mCherry were used as monomeric standards, while TdTomato was used as a dimeric standard. (b) Size-exclusion chromatography of EGFP, CyRFP1, and CyRFP1 A161K. (c) Absorbance of mCyRFP1. (d) Excitation and emission spectra of CyOFP1, CyRFP1, and mCyRFP1. (e) pH dependence of mCyRFP1 fluorescence, demonstrating a pKa of 5.64. (f) Photobleaching kinetics of purified cyan-excitable orange-red fluorescent proteins under illumination by a 120-W metal-halide arc lamp through a 490/20-nm excitation filter. The time-axis was adjusted for each fluorophore to simulate excitation conditions producing 1000 photons per s per molecule. Lighter shading on CyOFP1, CyRFP1, and mCyRFP1 lines represents standard deviation of four measurements.
Supplementary Figure 2 Mono-exponential lifetime decay of mCyRFP1.
Representative raw photon counting curves of single HEK cells expressing mEGFP (a), ECFP (b), CyRFP1(c) and mCyRFP1 (d). For each decay curve, a mono- and bi- exponential fit and weighted residuals are shown.
Supplementary Figure 3 CyRFP1 and mCyRFP1 characterization in mammalian cells.
(a) Relative brightness of HeLa cells expressing cyan-excitable orange-red FPs. Orange-red FPs fused to the CAAX membrane-localization signal were coexpressed with EGFP. The ratio of orange-red to green fluorescence was calculated, and charted relative to the value for CyOFP1. Error bars are standard deviation of duplicate measurements. (b) Photostability of cyan-excitable orange-red fluorescent proteins in HeLa cells under illumination by a 120-W metal-halide arc lamp through a 485/10-nm excitation filter at 100% neutral density. Emission was normalized to initial output, and no adjustments for per-molecule brightness were performed. Lighter shading represents standard deviation of four measurements.
Supplementary Figure 4 Performance of mCyRFP1 in protein fusions.
(a) Fluorescence images of mCyRFP1-10aa-H2B (human, nucleosomes) in (i) interphase, (ii) prophase, (iii) metaphase, (iv) anaphase. Scale bar, 10 μm. (b) Fluorescence images of HeLa cells expressing mCyRFP1 fused to various domains. For each fusion, the linker amino acid (aa) length is indicated in between the two domains and the origin of the fusion partner and its normal subcellular location are indicated in parentheses. (i) mCyRFP1-7aa-actin (human, actin cytoskeleton), (ii) mCyRFP-2aa-tubulin (human, microtubules), (iii) paxillin-22aa-mCyRFP1 (chicken, focal adhesions), (iv) PDHA-10aa-mCyRFP1 (human; mitochondrial pyruvate dehydrogenase), (v) mCyRFP1-10aa-lamin B1 (human, nuclear envelope), (vi) Calnexin-14aa-mCyRFP1 (human, endoplasmic reticulum), (vii) mannosidaseII-10aa-mCyRFP1 (mouse, Golgi complex), (viii) connexin43-7aa-mCyRFP1 (rat, cell-cell adhesion junctions), (ix) mCyRFP1-2aa-CAAX. Scale bar, 10 μm. (c) Performance of mCyRFP1 compared to mEGFP and mCardinal on CytERM monomericity assay. Error bars represent standard deviation of measurements from > 150 cells in each of three separate experiments.
Supplementary Figure 5 Two-photon performance of CyRFP1 and mCyRFP1.
(a) 2p excitation spectra of purified mEGFP, mMaroon1, CyRFP1, mCyRFP1 (1 mg/ml). Fluorescence was collected through the same PMT after a short-pass wavelength filter (Chroma, sp670). Raw fluorescence values for each FP were normalized to square root of measured power at each wavelength. Average of 2 measurements. (b) 2p excitation spectra normalized to their peaks. Same data as presented in (a). (c) Quantification of bleed-through of CyRFP1, mCyRFP1 or CyOFP1 into the green channel (490-550 nm band-pass filter) in HEK-293 cells. The ratios of photon counts measured in the green channel relative to that in the red channel are presented. Number of cells are 60, 37 and 58 for CyRFP1, mCyRFP1 and CyOFP1, respectively. (d) Fluorescence intensity of dendritic spines in CA1 pyramidal neurons expressing mEGFP together with either mCyRFP1 or CyRFP1 in organotypic hippocampal slices of mice. Intensity was normalized to the average of the first 4 images. Images were collected every 10-15 seconds with average excitation power of 1.5-2 mW under the objective. Data shown from 5 neurons/ 18 spines for mCyRFP1, 5 neurons / 21 spines for CyRFP1 and total of 10 neurons / 39 spines for mEGFP.
Supplementary Figure 6 Comparison of aggregation of RFPs.
(a) Representative images of the neuronal somata in organotypic hippocampal slices expressing various RFPs together with mEGFP. Images were acquired 7-10 days following transfection. Scale bar- 10 μm. (b) The ratios of the number of cells displaying RFP aggregation to the total number of cells imaged.
Supplementary Figure 7 Characterization of the mCyRFP1-mMaroon1 FRET pair for FLIM.
(a) Comparison of the FRET efficiency for mCyRFP1 fused to mNeptune2, mCardinal and Maroon, expressed in HEK cells. FRET efficiency of the population undergoing FRET was 46.0±0.01%, 52.7±0.005%, 62.3±0.002%, respectively. Quantification of 28, 22 and 34 fields of view. (b) Comparison of folding efficiency of mNeptune2, mCardinal and Maroon fused to mCyRFP1 in HEK cells. Folding efficiency was 42.0±0.01% (n=103 cells), 47.3±0.01% (n=138 cells) and 69.2±0.003% (n=160 cells), respectively. (c) Comparison of fluorescence lifetime of fluorophores in HEK293 cells. Green: fluorescence lifetime of mEGFP expressed alone (2.62±0.004ns, n=65 cells) is compared with that of mEGFP expressed together with mCyRFP1 (2.62±0.004ns, n=66 cells) or mCyRFP1-Maroon1 fusion (2.58±0.006ns, n=51 cells). Red: fluorescence lifetime of mCyRFP1 expressed alone (3.58±0.002ns, n=95 cells) is compared with that expressed together with mEGFP (3.56±0.005ns, n=106 cells), with mEGFP-sREACh fusion (3.49±0.007ns, n=67 cells), with mEGFP-CaMKII-dimVenus (3.52±0.002ns, n=153 cells) or with mMaroon1 (3.51±0.005ns, n=81 cells). (d) Comparison of binding fraction for FRET-FLIM pairs expressed in HEK cells, for mEGFP-sREACh pair alone (0.75±0.003, n=139 cells), with mCyRFP1 (0.73±0.004, n=111 cells) or with mCyRFP1-Maroon1 (0.74±0.004, n=78 cells). For mCyRFP1-Maroon pair alone (0.69±0.003, n=160 cells), with mEGFP (0.65±0.005, n=49 cells), with mEGFP-sREACh (0.66±0.008, n=73 cells) or with mEGFP-CaMKII-dimVenus (0.70±0.003, n=104 cells).
Supplementary Figure 8 Control experiments for RhoA/CaMKII imaging during spine structural plasticity.
(a) Fluorescence lifetime of dimVenus alone or the dimVenus-mMaroon1 fusion expressed in HEK cells. Averaged fluorescence lifetime is 0.33±0.015 ns (55 cells) and 0.35±0.005 ns (70 cells) for dimVenus and dimVenus-mMaroon, respectively. (b-d) Fluorescent lifetime changes of RhoA and CaMKII sensors in stimulated spines and nearby dendrites during dual FLIM imaging under the following conditions: (b) Green-Camuiα /RhoA-CyRM without extracellular Ca+2 (replaced with 4mM Mg+2), (c) mEGFP-dimVenus (no CaMKII)/RhoA-CyRM, and (d) Green-Camuiα /CyRFP-RhoA in combination with mMaroon-PAK3(60-113)/S74A/F84A-mMaroon (Cdc42-CyRM acceptor). Number of spines/cells are 10/8, 7/6 and 8/6 spines/cells respectively. Line is mean value across experiments and shaded colors represent SEM.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1–4 and Supplementary Note. (PDF 1439 kb)
CyRFP1/GCaMP6s in vivo imaging
A movie showing a dendritic segment of a layer 2/3 pyramidal neuron expressing GCaMP6s (Cyan) and CyRFP1 (Red) during spontaneous calcium dynamics in the motor cortex of a lightly anesthetized mouse. Raw fluorescence intensity was adjusted for each channel and filtered using Gaussian blur, radius of 1. Speed x2 from real time (acquired at 16 Hz). (AVI 57414 kb)
Rights and permissions
About this article
Cite this article
Laviv, T., Kim, B., Chu, J. et al. Simultaneous dual-color fluorescence lifetime imaging with novel red-shifted fluorescent proteins. Nat Methods 13, 989–992 (2016). https://doi.org/10.1038/nmeth.4046
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4046
This article is cited by
-
Fluorescent sensors for imaging of interstitial calcium
Nature Communications (2023)
-
An Algorithm Based on a Cable-Nernst Planck Model Predicting Synaptic Activity throughout the Dendritic Arbor with Micron Specificity
Neuroinformatics (2023)
-
Cloning and structural basis of fluorescent protein color variants from identical species of sea anemone, Diadumene lineata
Photochemical & Photobiological Sciences (2023)
-
A red fluorescent protein with improved monomericity enables ratiometric voltage imaging with ASAP3
Scientific Reports (2022)
-
Two-photon excitation fluorescent spectral and decay properties of retrograde neuronal tracer Fluoro-Gold
Scientific Reports (2021)