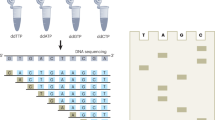
Abstract
Large-scale mutagenesis of target DNA sequences allows researchers to comprehensively assess the effects of single-nucleotide changes. Here we demonstrate the construction of a systematic allelic series (SAS) using massively parallel single-nucleotide mutagenesis with reversibly terminated deoxyinosine triphosphates (rtITP). We created a mutational library containing every possible single-nucleotide mutation surrounding the active site of the TEM-1 β-lactamase gene. When combined with high-throughput functional assays, SAS mutational libraries can expedite the functional assessment of genetic variation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Findlay, G.M., Boyle, E.A., Hause, R.J., Klein, J.C. & Shendure, J. Nature 513, 120–123 (2014).
Kitzman, J.O., Starita, L.M., Lo, R.S., Fields, S. & Shendure, J. Nat. Methods 12, 203–206 (2015).
Patwardhan, R.P. et al. Nat. Biotechnol. 30, 265–270 (2012).
Patwardhan, R.P. et al. Nat. Biotechnol. 27, 1173–1175 (2009).
Starita, L.M. et al. Genetics 200, 413–422 (2015).
Smith, R.P. et al. Nat. Genet. 45, 1021–1028 (2013).
Cirino, P.C., Mayer, K.M. & Umeno, D. Methods Mol. Biol. 231, 3–9 (2003).
Copp, J.N., Hanson-Manful, P., Ackerley, D.F. & Patrick, W.M. Methods Mol. Biol. 1179, 3–22 (2014).
McCullum, E.O., Williams, B.A., Zhang, J. & Chaput, J.C. Methods Mol. Biol. 634, 103–109 (2010).
Gao, Y. et al. Wei Sheng Wu Xue Bao 54, 97–103 (2014).
Stiffler, M.A., Hekstra, D.R. & Ranganathan, R. Cell 160, 882–892 (2015).
Acknowledgements
We thank C. Cruchaga and J. Budde and members of the Gurnett/Dobbs Lab for helpful discussion. This work was supported by a postdoctoral research fellowship (84291-STL) from the Shriners Hospital for Children (G.H.); US National Institutes of Health (NIH) grants (R01AR067715-01) (C.A.G. and M.B.D.) and (R01NS076993) (R.D.M.); and Shriners Hospital for Children research grant (85200-STL) (C.A.G. and M.B.D.)
Author information
Authors and Affiliations
Contributions
G.H., D.A., R.D.M., M.B.D. and C.A.G. designed the study and wrote the manuscript. G.H. and K.M. performed experiments. All authors contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Washington University in St. Louis has filed a provisional patent application on this method, with G.H., D.A., C.A.G. and M.B.D. as inventors.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1 and 2, and Supplementary Note. (PDF 915 kb)
Source data
Rights and permissions
About this article
Cite this article
Haller, G., Alvarado, D., McCall, K. et al. Massively parallel single-nucleotide mutagenesis using reversibly terminated inosine. Nat Methods 13, 923–924 (2016). https://doi.org/10.1038/nmeth.4015
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4015
This article is cited by
-
The evolution of substrate discrimination in macrolide antibiotic resistance enzymes
Nature Communications (2018)
-
Boosting the efficiency of site-saturation mutagenesis for a difficult-to-randomize gene by a two-step PCR strategy
Applied Microbiology and Biotechnology (2018)