Abstract
Mass spectrometry (MS) is the main technology used in proteomics approaches. However, on average, 75% of spectra analyzed in an MS experiment remain unidentified. We propose to use spectrum clustering at a large scale to shed light on these unidentified spectra. The Proteomics Identifications (PRIDE) Database Archive is one of the largest MS proteomics public data repositories worldwide. By clustering all tandem MS spectra publicly available in the PRIDE Archive, coming from hundreds of data sets, we were able to consistently characterize spectra into three distinct groups: (1) incorrectly identified, (2) correctly identified but below the set scoring threshold, and (3) truly unidentified. Using multiple complementary analysis approaches, we were able to identify ∼20% of the consistently unidentified spectra. The complete spectrum-clustering results are available through the new version of the PRIDE Cluster resource (http://www.ebi.ac.uk/pride/cluster). This resource is intended, among other aims, to encourage and simplify further investigation into these unidentified spectra.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Chick, J.M. et al. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat. Biotechnol. 33, 743–749 (2015).
Eng, J.K., McCormack, A.L. & Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Craig, R. & Beavis, R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Frank, A. & Pevzner, P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964–973 (2005).
Tabb, D.L., Ma, Z.Q., Martin, D.B., Ham, A.J. & Chambers, M.C. DirecTag: accurate sequence tags from peptide MS/MS through statistical scoring. J. Proteome Res. 7, 3838–3846 (2008).
Lam, H. et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 (2007).
Ma, C.W. & Lam, H. Hunting for unexpected post-translational modifications by spectral library searching with tier-wise scoring. J. Proteome Res. 13, 2262–2271 (2014).
Vizcaíno, J.A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 (2016).
Vizcaíno, J.A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014).
Griss, J., Foster, J.M., Hermjakob, H. & Vizcaíno, J.A. PRIDE Cluster: building a consensus of proteomics data. Nat. Methods 10, 95–96 (2013).
Yao, Q. et al. Design and development of a medical big data processing system based on Hadoop. J. Med. Syst. 39, 23 (2015).
Hodor, P., Chawla, A., Clark, A. & Neal, L. cl-dash: rapid configuration and deployment of Hadoop clusters for bioinformatics research in the cloud. Bioinformatics 32, 301–303 (2016).
Dasari, S. et al. Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment. J. Proteome Res. 11, 1686–1695 (2012).
Frank, A.M. et al. Spectral archives: extending spectral libraries to analyze both identified and unidentified spectra. Nat. Methods 8, 587–591 (2011).
The, M. & Kall, L. MaRaCluster: a fragment rarity metric for clustering fragment spectra in shotgun proteomics. J. Proteome Res. 15, 713–720 (2016).
Ternent, T. et al. How to submit MS proteomics data to ProteomeXchange via the PRIDE database. Proteomics 14, 2233–2241 (2014).
Desiere, F. et al. The PeptideAtlas project. Nucleic Acids Res. 34, D655–D658 (2006).
Craig, R., Cortens, J.P. & Beavis, R.C. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 (2004).
Omenn, G.S. et al. Metrics for the Human Proteome Project 2015: progress on the human proteome and guidelines for high-confidence protein identification. J. Proteome Res. 14, 3452–3460 (2015).
Hu, Y. & Lam, H. Expanding tandem mass spectral libraries of phosphorylated peptides: advances and applications. J. Proteome Res. 12, 5971–5977 (2013).
Liu, Y. et al. Chromosome-8-coded proteome of Chinese Chromosome Proteome Data set (CCPD) 2.0 with partial immunohistochemical verifications. J. Proteome Res. 13, 126–136 (2014).
Tsai, C.F. et al. Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Anal. Chem. 86, 685–693 (2014).
Ye, X. & Li, L. Macroporous reversed-phase separation of proteins combined with reversed-phase separation of phosphopeptides and tandem mass spectrometry for profiling the phosphoproteome of MDA-MB-231 cells. Electrophoresis 35, 3479–3486 (2014).
Mancuso, F., Bunkenborg, J., Wierer, M. & Molina, H. Data extraction from proteomics raw data: an evaluation of nine tandem MS tools using a large Orbitrap data set. J. Proteomics 75, 5293–5303 (2012).
Raijmakers, R., Kraiczek, K., de Jong, A.P., Mohammed, S. & Heck, A.J. Exploring the human leukocyte phosphoproteome using a microfluidic reversed-phase-TiO2-reversed-phase high-performance liquid chromatography phosphochip coupled to a quadrupole time-of-flight mass spectrometer. Anal. Chem. 82, 824–832 (2010).
Casado, P. et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci. Signal. 6, rs6 (2013).
Menschaert, G. et al. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12, 1780–1790 (2013).
Casado, P., Bilanges, B., Rajeeve, V., Vanhaesebroeck, B. & Cutillas, P.R. Environmental stress affects the activity of metabolic and growth factor signaling networks and induces autophagy markers in MCF7 breast cancer cells. Mol. Cell. Proteomics 13, 836–848 (2014).
Collins, M.O., Wright, J.C., Jones, M., Rayner, J.C. & Choudhary, J.S. Confident and sensitive phosphoproteomics using combinations of collision induced dissociation and electron transfer dissociation. J. Proteomics 103, 1–14 (2014).
van Gestel, R.A. et al. Quantitative erythrocyte membrane proteome analysis with Blue-native/SDS PAGE. J. Proteomics 73, 456–465 (2010).
Sleno, L. The use of mass defect in modern mass spectrometry. J. Mass Spectrometry 47, 226–236 (2012).
Sturm, M. et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163 (2008).
Wang, J., Pérez-Santiago, J., Katz, J.E., Mallick, P. & Bandeira, N. Peptide identification from mixture tandem mass spectra. Mol. Cell. Proteomics 9, 1476–1485 (2010).
Schittmayer, M., Fritz, K., Liesinger, L., Griss, J. & Birner-Gruenberger, R. Cleaning out the litterbox of proteomic scientists' favorite pet: optimized data analysis avoiding trypsin artifacts. J. Proteome Res. 15, 1222–1229 (2016).
Lam, H. Spectral archives: a vision for future proteomics data repositories. Nat. Methods 8, 546–548 (2011).
Mosteller, F., Winsor, C.P. & Fisher, C.H. Questions and Answers. Am. Stat. 2, 18–19 (1948).
Mi, H., Muruganujan, A. & Thomas, P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 (2013).
Acknowledgements
This work was supported by the Vienna Science and Technology Fund (WWTF, grant LS11-045; grant was awarded to S.N. Wagner (Medical University of Vienna, Division of Immunology, Allergy and Infectious Diseases) and used to fund J.G.), the Wellcome Trust (grant WT101477MA to H.H. and J.A.V.), the BBSRC ('PROCESS' grant BB/K01997X/1 to H.H. and J.A.V., 'Quantitative Proteomics' grant BB/I00095X/1 to H.H.), the Deutsche Forschungsgemeinschaft (grant SFB685/B1 to O.K.), and the BMBF (grant 01ZX1301F to O.K.). We would like to acknowledge the attendees of the Midwinter Proteomics Bioinformatics Seminar 2015 at Semmering (Austria) and the Bioinformatics Hub at the HUPO conference 2015 at Vancouver (Canada), who provided valuable feedback on the data analysis. Finally, we want to acknowledge M. The and L. Käll for their support during the benchmarking of their MaRaCluster algorithm.
Author information
Authors and Affiliations
Contributions
J.G. developed the clustering algorithm, ran the experiments, and performed the data analysis. D.L.T. contributed to the development of the probabilistic scoring approach. Y.P.-R. contributed to the data analysis. J.G. and R.W. developed the Java APIs for the spectrum-clustering-analysis pipeline. S.L., R.W., and J.G. developed the Hadoop implementation. J.A.D., N.d.-T., Y.P.-R., and R.W. created the web interface and the API of the PRIDE Cluster resource. M.R., M.W., and O.K. performed the metabolite search. J.G., R.W., H.H., and J.A.V. supervised the project. J.G. and J.A.V. wrote the manuscript, with contributions from the rest of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Relative Proportion of Unidentified Spectra in datasets submitted to PRIDE Archive
Box plots representing the relative proportion of unidentified spectra in the PRoteomics IDEntifications (PRIDE) Archive database. Overall, 75% of spectra in datasets submitted to PRIDE Archive are unidentified. Submitted datasets without identified spectra as well as datasets that only contained identified spectra were excluded from this calculation. Submissions to PRIDE (first box plot) represent those datasets submitted until mid-2012. Submissions to ProteomeXchange (second box plot) represent datasets submitted afterwards, once the ProteomeXchange data workflows were started. For the latter, only “complete” submissions are considered.
Supplementary Figure 2 Peptide Evidence per Proteomics Repository
Venn diagrams demonstrating that PRIDE Cluster based reliable peptide identifications provide additional MS/MS evidence for peptides not found in the other two other major MS-based data repositories (PeptideAtlas and GPMDB). Data are shown for (a) human, (b) mouse, (c) Arabidopsis thaliana, and (d) rat.
Supplementary Figure 3 PRIDE Cluster Provides MS/MS Evidence for Proteins without Experimental Evidence Annotated
PRIDE Cluster based validated peptide spectrum matches provide experimental evidence for the existence of a considerable number of proteins for which there is no experimental evidence at the protein level (PE=1), in UniProt. The present plot present the list of proteins that can be identified with at least 2 unique peptides with at least 9 amino acids, as was agreed in the latest guidelines of the Human Proteome Project. The categories represented are: only evidence on transcript level (PE=2), proteins inferred from homology (PE=3), and predicted proteins (PE=4) in the human UniProtKB/SwissProt (release 2016-03) database.
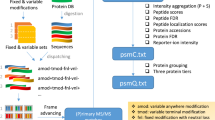
Supplementary Figure 4 Overall workflow representing the “identification pipeline”.
Workflow representing the “identification pipeline” used to identify originally submitted spectra of incorrectly identified and unidentified clusters.
Supplementary Figure 5 Overall workflow representing the PRIDE Cluster analysis process.
Flow chart summarizing all analyses steps performed during the analyses of the spectrum clustering results, as described in the main manuscript.
Supplementary Figure 6 Open modification search of unidentified mouse clusters.
Summary of results for the analysis of mouse clusters containing only unidentified spectra. The vast majority of delta masses observed in the open modification search were between -2 and +4 Da (top left panel). After adjusting the y-axis, several other delta masses were observed at a high frequency (top right panel). When limiting these delta masses to only masses that were observed at least for ten different clusters, the vast majority of delta masses could be mapped to known PTMs, as well as to one potential amino acid substitution (lower panel). For the complete list of the found delta masses see Supplementary Table 4.
Supplementary Figure 7 Open modification search of unidentified Arabidopsis thaliana clusters.
Summary of results for the analysis of Arabidopsis thaliana clusters containing only unidentified spectra. In contrast to the human and mouse data, consensus spectra of unidentified A. thaliana clusters were searched against the PRIDE Cluster spectral library for A. thaliana (version 2015-04). Again, most delta masses were found between -1 and 1 Da (top left panel). After adjusting the y-axis several other delta masses were observed (top right panel). When limiting these delta masses to only masses that were observed at least for five different clusters, three known PTMs could be identified even taking into account that the spectral library used in this search was derived from the same dataset (lower panel). For the complete list of the found delta masses see Supplementary Table 4.
Supplementary Figure 8 Identification of unidentified mouse clusters
Overview of the results of the analysis of clusters containing only unidentified spectra from mouse. (a) Venn diagram representing that 122 (15%) of the large unidentified mouse clusters were identified using SpectraST, X!Tandem and PepNovo. (b) In contrast to the results in human data, around 50% of identified proteins could not be classified as albumin, keratin, trypsin or haemoglobin. (c) Similarly to the human data, only trypsin peptides were commonly modified (e.g. dimethylated, the center line marks the median, edges the first and third quartile, whiskers extend to +/-1.58 times the inter-quartile ratio divided by the square root of the number of observations, single points denote measurements outside this range).
Supplementary Figure 9 Identification of unidentified Arabidopsis thaliana clusters
Overview of the results of the analysis of clusters containing only unidentified spectra from A. thaliana data. (a) Venn diagram representing that 50 (9%) of the large unidentified A. thaliana clusters were identified using SpectraST, X!Tandem and PepNovo. (b) In contrast to mouse and human data, no haemoglobin associated proteins were identified. Similarly to the identified proteins in the mouse dataset, most proteins could not be classified as albumin, keratin, or trypsin. All identified peptides corresponding to albumin were matches against bovine albumin and most likely experimental contaminants. (c) Similarly to the human and mouse data, trypsin peptides were commonly modified (e.g. dimethylated). Additionally, in this case the majority of albumin peptides were also modified (the center line marks the median, edges the first and third quartile, whiskers extend to +/-1.58 times the inter-quartile ratio divided by the square root of the number of observations, single points denote measurements outside this range).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Notes 1–8 (PDF 3504 kb)
Supplementary Table 1
List of processed human PRIDE Archive submissions as part of the test dataset (XLS 94 kb)
Supplementary Table 2
List of analysed phosphorylation studies submitted to PRIDE Archive. (XLS 123 kb)
Supplementary Table 3
List of identified phosphorylated peptides identified in the three examples presented in the manuscript. (XLS 79 kb)
Supplementary Table 4
List of commonly observed mass deltas when processing unidentified clusters using an open modification search. Results are given for human, mouse and Arabidopsis. (XLS 28 kb)
Rights and permissions
About this article
Cite this article
Griss, J., Perez-Riverol, Y., Lewis, S. et al. Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nat Methods 13, 651–656 (2016). https://doi.org/10.1038/nmeth.3902
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3902
This article is cited by
-
Fast alignment of mass spectra in large proteomics datasets, capturing dissimilarities arising from multiple complex modifications of peptides
BMC Bioinformatics (2023)
-
Progressive search in tandem mass spectrometry
BMC Bioinformatics (2023)
-
Proteomic analyses reveal cystatin c is a promising biomarker for evaluation of systemic lupus erythematosus
Clinical Proteomics (2023)
-
Spectroscape enables real-time query and visualization of a spectral archive in proteomics
Nature Communications (2023)
-
A joint proteomic and genomic investigation provides insights into the mechanism of calcification in coccolithophores
Nature Communications (2023)