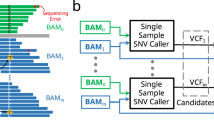
Abstract
Current variant callers are not suitable for single-cell DNA sequencing, as they do not account for allelic dropout, false-positive errors and coverage nonuniformity. We developed Monovar (https://bitbucket.org/hamimzafar/monovar), a statistical method for detecting and genotyping single-nucleotide variants in single-cell data. Monovar exhibited superior performance over standard algorithms on benchmarks and in identifying driver mutations and delineating clonal substructure in three different human tumor data sets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Navin, N.E. Genome Res. 25, 1499–1507 (2015).
Wang, Y. & Navin, N.E. Mol. Cell 58, 598–609 (2015).
Navin, N.E. Genome Biol. 15, 452 (2014).
Navin, N. et al. Nature 472, 90–94 (2011).
Garvin, T. et al. Nat. Methods 12, 1058–1060 (2015).
Stegle, O., Teichmann, S.A. & Marioni, J.C. Nat. Rev. Genet. 16, 133–145 (2015).
Brennecke, P. et al. Nat. Methods 10, 1093–1095 (2013).
Wang, Y. et al. Nature 512, 155–160 (2014).
Zong, C., Lu, S., Chapman, A.R. & Xie, X.S. Science 338, 1622–1626 (2012).
Wang, J., Fan, H.C., Behr, B. & Quake, S.R. Cell 150, 402–412 (2012).
McKenna, A. et al. Genome Res. 20, 1297–1303 (2010).
Li, H. et al. Bioinformatics 25, 2078–2079 (2009).
Li, R. et al. Bioinformatics 25, 1966–1967 (2009).
Goya, R. et al. Bioinformatics 26, 730–736 (2010).
Koboldt, D.C. et al. Genome Res. 22, 568–576 (2012).
Leung, M.L., Wang, Y., Waters, J. & Navin, N.E. Genome Biol. 16, 55 (2015).
Li, Y. et al. GigaScience 1, 12 (2012).
Gawad, C., Koh, W. & Quake, S.R. Proc. Natl. Acad. Sci. USA 111, 17947–17952 (2014).
Klein, A.M. et al. Cell 161, 1187–1201 (2015).
Macosko, E.Z. et al. Cell 161, 1202–1214 (2015).
Li, H. Bioinformatics 27, 2987–2993 (2011).
Hodgkinson, A. & Eyre-Walker, A. Genetics 184, 233–241 (2010).
You, N. et al. Bioinformatics 28, 643–650 (2012).
Le, S.Q. & Durbin, R. Genome Res. 21, 952–960 (2011).
Nielsen, R., Korneliussen, T., Albrechtsen, A., Li, Y. & Wang, J. PLoS ONE 7, e37558 (2012).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
Wang, K., Li, M. & Hakonarson, H. Nucleic Acids Res. 38, e164 (2010).
Quinlan, A.R. & Hall, I.M. Bioinformatics 26, 841–842 (2010).
Forbes, S.A. et al. Nucleic Acids Res. 43, D805–D811 (2015).
Futreal, P.A. et al. Nat. Rev. Cancer 4, 177–183 (2004).
Adzhubei, I., Jordan, D.M. & Sunyaev, S.R. Curr. Protoc. Hum. Genet. Chapter 7, Unit 7.20 (2013).
Ng, P.C. & Henikoff, S. Nucleic Acids Res. 31, 3812–3814 (2003).
Acknowledgements
This work was supported by grants to N.N. from the Lefkofsky Foundation, NCI (RO1CA169244-01), NIH (R21CA174397), Agilent University Relations, and MD Anderson Knowledge Gap and Center for Genetics & Genomics. N.N. is a Damon Runyon-Rachleff Innovator (DRR-25-13), ACS Research Scholar, T.C. Hsu Endowed Scholar and Sabin Fellow. K.C. is a Sabin Fellow and was supported by an NCI grant (RO1CA172652). The study was supported by the Bosarge, Chapman and Dell Foundations and NCI (CA016672). The authors thank W. Zhou.
Author information
Authors and Affiliations
Contributions
H.Z. was involved in all aspects. Y.W. analyzed the data. L.N. developed the algorithm. N.N. analyzed the data and wrote the manuscript. K.C. analyzed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Comparison of Monovar to Standard Variant Calling Methods using Isogenic Cell Line Data
Monovar, GATK UnifiedGenotyper, GATK HaplotypeCaller and Samtools were compared using single cell exome sequencing data generated from a normal isogenic fibroblast cell line in terms of SNV detection (a) Precision versus Detection Efficiency (Recall) and (b) SNV transition and transversion spectrum for FP errors.
Supplementary Figure 2 Performance of Monovar using in Silico Down-Sampled Coverage Depth Data
Monovar and GATK HaplotypeCaller were compared in terms of (a) Precision and (b) Detection Efficiency (Recall), respectively at 40×, 30×, 20× and 10× sequencing depths, acquired via down-sampling the SKN2 SCS data.
Supplementary Figure 3 Performance of Monovar for Detecting Sub-clonal SNVs in Admixture Samples
The SNV detection (a) Precision and (b) Detection Efficiency of Monovar were measured by comparing SNVs detected from a set of datasets, created by in silico intermixing of variable numbers of SKN2 and 12 TNBC cells, with SNVs detected from SKN2 bulk sequencing data.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–6 and Supplementary Note 1 (PDF 2042 kb)
Supplementary Software
Monovar code and accessory files. (ZIP 28383 kb)
Rights and permissions
About this article
Cite this article
Zafar, H., Wang, Y., Nakhleh, L. et al. Monovar: single-nucleotide variant detection in single cells. Nat Methods 13, 505–507 (2016). https://doi.org/10.1038/nmeth.3835
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3835
This article is cited by
-
SCSilicon: a tool for synthetic single-cell DNA sequencing data generation
BMC Genomics (2022)
-
Single-cell genome sequencing of human neurons identifies somatic point mutation and indel enrichment in regulatory elements
Nature Genetics (2022)
-
MQuad enables clonal substructure discovery using single cell mitochondrial variants
Nature Communications (2022)
-
Bisulfite-free epigenomics and genomics of single cells through methylation-sensitive restriction
Communications Biology (2021)
-
Clinical and therapeutic relevance of cancer-associated fibroblasts
Nature Reviews Clinical Oncology (2021)