Abstract
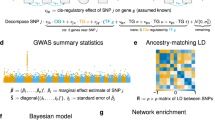
Mapping perturbed molecular circuits that underlie complex diseases remains a great challenge. We developed a comprehensive resource of 394 cell type– and tissue-specific gene regulatory networks for human, each specifying the genome-wide connectivity among transcription factors, enhancers, promoters and genes. Integration with 37 genome-wide association studies (GWASs) showed that disease-associated genetic variants—including variants that do not reach genome-wide significance—often perturb regulatory modules that are highly specific to disease-relevant cell types or tissues. Our resource opens the door to systematic analysis of regulatory programs across hundreds of human cell types and tissues (http://regulatorycircuits.org).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Marstrand, T.T. & Storey, J.D. Identifying and mapping cell-type-specific chromatin programming of gene expression. Proc. Natl. Acad. Sci. USA 111, E645–E654 (2014).
Pers, T.H. et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 6, 5890 (2015).
Roy, S. et al. A predictive modeling approach for cell line-specific long-range regulatory interactions. Nucleic Acids Res. 43, 8694–8712 (2015).
Ward, L.D. & Kellis, M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 30, 1095–1106 (2012).
Parker, S.C.J. et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA 110, 17921–17926 (2013).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Faye, L.L., Machiela, M.J., Kraft, P., Bull, S.B. & Sun, L. Re-ranking sequencing variants in the post-GWAS era for accurate causal variant identification. PLoS Genet. 9, e1003609 (2013).
Pickrell, J.K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94, 559–573 (2014).
Pasquali, L. et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 46, 136–143 (2014).
Navlakha, S. & Kingsford, C. The power of protein interaction networks for associating genes with diseases. Bioinformatics 26, 1057–1063 (2010).
Rossin, E.J. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Lee, I., Blom, U.M., Wang, P.I., Shim, J.E. & Marcotte, E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 21, 1109–1121 (2011).
Mäkinen, V.-P. et al. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 10, e1004502 (2014).
Pierson, E., Koller, D., Battle, A., Mostafavi, S. & the GTEx Consortium. Sharing and specificity of co-expression networks across 35 human tissues. PLoS Comput. Biol. 11, e1004220 (2015).
Greene, C.S. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015).
Gerstein, M.B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012).
Marbach, D. et al. Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res. 22, 1334–1349 (2012).
Karczewski, K.J., Snyder, M., Altman, R.B. & Tatonetti, N.P. Coherent functional modules improve transcription factor target identification, cooperativity prediction, and disease association. PLoS Genet. 10, e1004122 (2014).
Boyer, L.A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005).
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012).
Chen, J.C. et al. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell 159, 402–414 (2014).
The FANTOM Consortium & the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
Kheradpour, P. et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 23, 800–811 (2013).
Kheradpour, P. & Kellis, M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42, 2976–2987 (2014).
Marbach, D. et al. Wisdom of crowds for robust gene network inference. Nat. Methods 9, 796–804 (2012).
The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Lamparter, D., Marbach, D., Rico, R., Kutalik, Z. & Bergmann, S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput. Biol. 12, e1004714 (2016).
Perez-Costas, E., Melendez-Ferro, M. & Roberts, R.C. Basal ganglia pathology in schizophrenia: dopamine connections and anomalies. J. Neurochem. 113, 287–302 (2010).
Garrett, A. & Chang, K. The role of the amygdala in bipolar disorder development. Dev. Psychopathol. 20, 1285–1296 (2008).
Cromer, W.E., Mathis, J.M., Granger, D.N., Chaitanya, G.V. & Alexander, J.S. Role of the endothelium in inflammatory bowel diseases. World J. Gastroenterol. 17, 578–593 (2011).
Swirski, F.K. et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325, 612–616 (2009).
Chichlowski, M., Westwood, G.S., Abraham, S.N. & Hale, L.P. Role of mast cells in inflammatory bowel disease and inflammation-associated colorectal neoplasia in IL-10-deficient mice. PLoS ONE 5, e12220 (2010).
Wright, H.L., Moots, R.J. & Edwards, S.W. The multifactorial role of neutrophils in rheumatoid arthritis. Nat. Rev. Rheumatol. 10, 593–601 (2014).
Iadecola, C. Neurovascular regulation in the normal brain and in Alzheimer's disease. Nat. Rev. Neurosci. 5, 347–360 (2004).
Hor, H. et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat. Genet. 42, 786–789 (2010).
Goodrich, J.K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Kheradpour, P., Stark, A., Roy, S. & Kellis, M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 17, 1919–1931 (2007).
Boyle, A.P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Marbach, D. et al. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl. Acad. Sci. USA 107, 6286–6291 (2010).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Ripke, S. et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45, 1150–1159 (2013).
Boraska, V. et al. A genome-wide association study of anorexia nervosa. Mol. Psychiatry 19, 1085–1094 (2014).
Lambert, J.-C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 45, 1452–1458 (2013).
Simón-Sánchez, J. et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 41, 1308–1312 (2009).
International Multiple Sclerosis Genetics Consortium. et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).
Anderson, C.A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43, 246–252 (2011).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Stahl, E.A. et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514 (2010).
Rauch, A. et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138, 1338–1345 (2010).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 (2011).
The International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011).
Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Prokopenko, I. et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 10, e1004235 (2014).
The DIAbetes Genetxzics Replication and Meta-analysis (DIAGRAM) Consortium. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Scott, R.A. et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 44, 991–1005 (2012).
Soranzo, N. et al. Common variants at 10 genomic loci influence hemoglobin A1C levels via glycemic and nonglycemic pathways. Diabetes 59, 3229–3239 (2010).
Dupuis, J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42, 105–116 (2010).
Strawbridge, R.J. et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2. Diabetes 60, 2624–2634 (2011).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Speliotes, E.K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
The AMD Gene Consortium. Seven new loci associated with age-related macular degeneration. Nat. Genet. 45, 433–439 (2013).
Estrada, K. et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44, 491–501 (2012).
Clarke, L. et al. The 1000 Genomes Project: data management and community access. Nat. Methods 9, 459–462 (2012).
Kondor, R.I. & Lafferty, J.D. Diffusion kernels on graphs and other discrete input spaces. in Proc. Nineteenth International Conference on Machine Learning 315–322 (Morgan Kaufmann, 2002).
Smola, A.J. & Kondor, R. Kernels and regularization on graphs. in Learning Theory and Kernel Machines (eds. Schölkopf, B. & Warmuth, M.K.) 144–158 (Springer Berlin Heidelberg, 2003).
Erten, S., Bebek, G., Ewing, R.M. & Koyutürk, M. DADA: degree-aware algorithms for network-based disease gene prioritization. BioData Min. 4, 19 (2011).
Lage, K. et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25, 309–316 (2007).
Maglott, D., Ostell, J., Pruitt, K.D. & Tatusova, T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 39, D52–D57 (2011).
Chatr-Aryamontri, A. et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 43, D470–D478 (2015).
Rolland, T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014).
Neph, S. et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell 150, 1274–1286 (2012).
Derry, J.M.J. et al. Developing predictive molecular maps of human disease through community-based modeling. Nat. Genet. 44, 127–130 (2012).
Acknowledgements
We thank P. Kheradpour (MIT) for providing the collection of curated TF binding motifs. This work was supported by the Swiss National Science Foundation (grant FN 310030_152724/1 to S.B. and grant FN 31003A-143914 to Z.K.), SystemsX.ch (grant SysGenetiX to S.B. and grant AgingX to Z.K.), the Swiss Institute of Bioinformatics (Z.K. and S.B.) and the Leenaards Foundation (Z.K.).
Author information
Authors and Affiliations
Contributions
D.M. designed the study, performed analyses and prepared the manuscript. D.L. performed gene scoring and phenotype-label permutation. D.M., D.L., G.Q., M.K., Z.K. and S.B. conceived methods, discussed the results and implications, and commented on the manuscript at all stages.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–47 (PDF 2364 kb)
Supplementary Table 1
Sample annotation (XLSX 108 kb)
Supplementary Table 2
ENCODE ChIP-seq experiments (XLSX 47 kb)
Supplementary Table 3
GTEx tissues (XLSX 42 kb)
Supplementary Table 4
Roadmap Epigenomics RNA-seq data (XLSX 46 kb)
Supplementary Table 5
GWAS compendium (XLSX 16 kb)
Supplementary Table 6
Links to external repositories (XLSX 53 kb)
Rights and permissions
About this article
Cite this article
Marbach, D., Lamparter, D., Quon, G. et al. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods 13, 366–370 (2016). https://doi.org/10.1038/nmeth.3799
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3799
This article is cited by
-
De-obstruction of bladder outlet in humans reverses organ remodelling by normalizing the expression of key transcription factors
BMC Urology (2024)
-
Optimized network based natural language processing approach to reveal disease comorbidities in COVID-19
Scientific Reports (2024)
-
Socioeconomic inequalities in early adulthood disrupt the immune transcriptomic landscape via upstream regulators
Scientific Reports (2024)
-
Prioritizing genes associated with brain disorders by leveraging enhancer-promoter interactions in diverse neural cells and tissues
Genome Medicine (2023)
-
Peripheral blood cellular dynamics of rheumatoid arthritis treatment informs about efficacy of response to disease modifying drugs
Scientific Reports (2023)