Abstract
The high volumes of data produced by state-of-the-art optical microscopes encumber research. We developed a method that reduces data size and processing time by orders of magnitude while disentangling signal by taking advantage of the laminar structure of many biological specimens. Our Image Surface Analysis Environment automatically constructs an atlas of 2D images for arbitrarily shaped, dynamic and possibly multilayered surfaces of interest. Built-in correction for cartographic distortion ensures that no information on the surface is lost, making the method suitable for quantitative analysis. We applied our approach to 4D imaging of a range of samples, including a Drosophila melanogaster embryo and a Danio rerio beating heart.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Krzic, U., Gunther, S., Saunders, T.E., Streichan, S.J. & Hufnagel, L. Nat. Methods 9, 730–733 (2012).
Tomer, R., Khairy, K., Amat, F. & Keller, P.J. Nat. Methods 9, 755–763 (2012).
Chen, B.-C. Science 346, 1257998 (2014).
Bosveld, F. et al. Science 336, 724–727 (2012).
Reynaud, E.G., Peychl, J., Huisken, J. & Tomancak, P. Nat. Methods 12, 30–34 (2015).
Alberts, B. et al. Molecular Biology of the Cell (Garland Science, 2014).
Gilbert, S.F. Developmental Biology (Sinauer Associates, 2014).
Amat, F. et al. Nat. Methods 11, 951–958 (2014).
Fowlkes, C.C. et al. Cell 133, 364–374 (2008).
Schmid, B. et al. Nat. Commun. 4, 2207 (2013).
Frankel, T. The Geometry of Physics: An Introduction (Cambridge Univ. Press, 2004).
Rolland-Lagan, A.-G., Bangham, J.A. & Coen, E. Nature 422, 161–163 (2003).
Clark, A.G., Dierkes, K. & Paluch, E.K. Biophys. J. 105, 570–580 (2013).
McDonald, D.M. & Choyke, P.L. Nat. Med. 9, 713–725 (2003).
Pitrone, P.G. et al. Nat. Methods 10, 598–599 (2013).
Sommer, C., Straehle, C., Kothe, U. & Hamprecht, F.A. Ilastik: interactive learning and segmentation toolkit. In Proc. 8th IEEE International Symposium on Biomedical Imaging (ISBI 2011) 230–233 (IEEE, 2011).
Mullen, P., Tong, Y., Alliez, P. & Desbrun, M. Spectral conformal parameterization. Comput. Graph. Forum 27, 1487–1494 (2008).
Klein, S., Staring, M., Murphy, K., Viergever, M.A. & Pluim, J.P.W. elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205 (2010).
Mickoleit, M. et al. High-resolution reconstruction of the beating zebrafish heart. Nat. Methods 11, 919–922 (2014).
Acknowledgements
This work was funded by the Gordon and Betty Moore Foundation (GBMF2919). We thank M. Rauzi for providing a MuVi-SPIM recording of the D. melanogaster embryo shown in Figure 1 and M. Albert for MuVI-SPIM fusion software. We also thank K. Irvine for providing the confocal data shown in Supplementary Figure 6. We thank B. Shraiman for discussions and participants in the 2013 summer session of the Santa Barbara Advanced School of Quantitative Biology for testing the software. We also thank B. Shraiman, D. Bilder and A. Warmflash for critical comments on the manuscript.
Author information
Authors and Affiliations
Contributions
I.H. and S.J.S. contributed equally to all aspects of this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
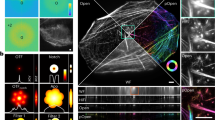
Supplementary Figure 1 Cylindrical coordinate lines in different charts.
The cylindrical coordinate system used to parameterize the embryo surface is shown in various charts and one 3D surface over a pullback of membrane labeled surface data just before gastrulation. Constant z lines in yellow, constant ϕ lines in cyan. (a) Cylinder map (b) Generalized sinusoidal map (c) 3D surface (d) Anterior ventral distance map (e) Posterior ventral distance map.
Supplementary Figure 2 Charts and metric strains.
Distortion in different charts. Like Supplementary Fig. 1 but including crosses displaying the distortion as measured by the strain. The two bars represent the major and minor axis of deformation, with compression in blue and extension in red. Red cross in corner represents positive unit strain. (a) Cylinder map (b) Generalized sinusoidal map (c) 3D surface (d) Anterior ventral distance map (e) Posterior ventral distance map.
Supplementary Figure 3 Performance gain from tissue cartography.
(a) Storage per time point in atlas versus raw data. Atlas normalized by number of surface coverings (number of times each point is contained in a map). Color codes for time dependence. Black: dynamic shapes require storage of atlas and geometric metadata for each time point; blue: static shapes with 10 time points only require storage of geometric metadata once, reducing the average storage per time point; red: as blue for 200 time points, further reducing average storage. Blue Square indicates average storage for the embryo data from Fig. 1, green diamond for heart data from Fig. 3. (b) Access and processing times for atlas and raw data in (a). Black: time required to load the data. Colors: typical processing times. Blue: Gaussian filter, magenta: erosion, red: theoretical data transfer at 5 MB/s of dynamic surface from (a). (c) Feature table of 3D, uncorrected 2D, and cartographic projections, comparing performance in routine tasks, data disentanglement, and feasibility of data post-processing methods.
Supplementary Figure 4 Triangulation of point cloud.
Workflow to construct a triangulation from a point cloud using Meshlab. (a) With a point cloud loaded, use filters tap and select show current filter step. (b) empty pop-up dialog, (c) filled with a sequence of filters. (d) Triangulation from applying the filter script by hitting apply script button.
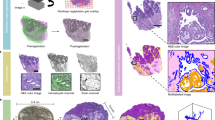
Supplementary Figure 6 Pullback versus z-projection in wing disc and notum.
Signal to noise ratio improvement of pullback relative to maximal intensity projection of E-Cad immunostaining of 3rd instar wing disc in (a), (b) and NRG-GFP expressing notum in 24 h old pupa (c), (d). (a), (c) maximal intensity projection (b), (d) pullback. Arrows highlight positions for comparison.
Supplementary Figure 7 Class diagram of ImSAnE.
UML-like class diagram of ImSAnE. Tabs are packages. Rectangles inside are classes, from top to bottom divided into: title, properties (variables) and methods (functions). Dashed line arrows denote dependence (requires). Solid line arrows denote inheritance. Solid lines denote a relationship.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Note 1 (PDF 3168 kb)
Supplementary Software
Image Surface Analysis Environment (ZIP 110809 kb)
Smoothly varying maps enable quantification of dynamic tissue geometry
Movie of valve region shown in Fig. 3c–g. Cyan, myocardium; red, endocardium. Select nuclei are pseudo–color-coded to indicate track identity. (MOV 457 kb)
Rights and permissions
About this article
Cite this article
Heemskerk, I., Streichan, S. Tissue cartography: compressing bio-image data by dimensional reduction. Nat Methods 12, 1139–1142 (2015). https://doi.org/10.1038/nmeth.3648
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3648
This article is cited by
-
VolumePeeler: a novel FIJI plugin for geometric tissue peeling to improve visualization and quantification of 3D image stacks
BMC Bioinformatics (2023)
-
Active cell divisions generate fourfold orientationally ordered phase in living tissue
Nature Physics (2023)
-
Movies tell us how tissues develop a 3D shape
Nature Methods (2023)
-
Mapping morphogenesis and mechanics in embryo models
Nature Methods (2023)
-
TubULAR: tracking in toto deformations of dynamic tissues via constrained maps
Nature Methods (2023)