Abstract
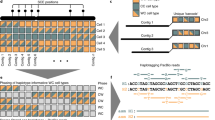
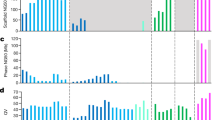
We present the first comprehensive analysis of a diploid human genome that combines single-molecule sequencing with single-molecule genome maps. Our hybrid assembly markedly improves upon the contiguity observed from traditional shotgun sequencing approaches, with scaffold N50 values approaching 30 Mb, and we identified complex structural variants (SVs) missed by other high-throughput approaches. Furthermore, by combining Illumina short-read data with long reads, we phased both single-nucleotide variants and SVs, generating haplotypes with over 99% consistency with previous trio-based studies. Our work shows that it is now possible to integrate single-molecule and high-throughput sequence data to generate de novo assembled genomes that approach reference quality.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Zook, J.M. et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 32, 246–251 (2014).
Lam, H.Y.K. et al. Performance comparison of whole-genome sequencing platforms. Nat. Biotechnol. 30, 78–82 (2012).
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007).
Istrail, S. et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl. Acad. Sci. USA 101, 1916–1921 (2004).
Gnerre, S. et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108, 1513–1518 (2011).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Human Genome Sequencing Consortium International. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Pang, A.W.C., Macdonald, J.R., Yuen, R.K.C., Hayes, V.M. & Scherer, S.W. Performance of high-throughput sequencing for the discovery of genetic variation across the complete size spectrum. G3 (Bethesda) 4, 63–65 (2014).
Schadt, E.E., Turner, S. & Kasarskis, A. A window into third generation sequencing. Hum. Mol. Genet. 19, R227–R240 (2010).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Mills, R.E. et al. Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65 (2011).
Ross, M.G. et al. Characterizing and measuring bias in sequence data. Genome Biol. 14, R51 (2013).
Rasko, D.A. et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717 (2011).
Bashir, A. et al. A hybrid approach for the automated finishing of bacterial genomes. Nat. Biotechnol. 30, 701–707 (2012).
Chin, C.-S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Ribeiro, F.J. et al. Finished bacterial genomes from shotgun sequence data. Genome Res. 22, 2270–2277 (2012).
Koren, S. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30, 693–700 (2012).
Huddleston, J. et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 24, 688–696 (2014).
Patel, A., Schwab, R., Liu, Y.-T. & Bafna, V. Amplification and thrifty single-molecule sequencing of recurrent somatic structural variations. Genome Res. 24, 318–328 (2014).
Hastie, A.R. et al. Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome. PLoS ONE 8, e55864 (2013).
Lam, E.T. et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 30, 771–776 (2012).
Salzberg, S.L. et al. GAGE: A critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22, 557–567 (2012).
Maccallum, I. et al. ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 10, R103 (2009).
Rozowsky, J. et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 7, 522 (2011).
Bansal, V., Halpern, A.L., Axelrod, N. & Bafna, V. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res. 18, 1336–1346 (2008).
Chaisson, M.J.P. et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2015).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Carter, A.B. et al. Genome-wide analysis of the human Alu Yb-lineage. Hum. Genomics 1, 167–178 (2004).
Myers, J.S. et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 71, 312–326 (2002).
Mason, C.E. et al. Location analysis for the estrogen receptor-alpha reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res. 38, 2355–2368 (2010).
Highnam, G. et al. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 41, e32 (2013).
Kamstrup, P.R. Lipoprotein(a) and ischemic heart disease–a causal association? A review. Atherosclerosis 211, 15–23 (2010).
Damert, A. et al. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 19, 1992–2008 (2009).
Xing, J. et al. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc. Natl. Acad. Sci. USA 103, 17608–17613 (2006).
Ejima, Y. & Yang, L. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum. Mol. Genet. 12, 1321–1328 (2003).
Ummat, A. & Bashir, A. Resolving complex tandem repeats with long reads. Bioinformatics 30, 3491–3498 (2014).
Myers, G. in Algorithms in Bioinformatics (eds. Brown, D. & Morgenstern, B.) 52–67 (Springer, 2014).
Berlin, K. et al. Assembling large genomes with single-molecule sequencing and locality sensitive hashing. bioRxiv doi:http://dx.doi.org/10.1101/008003 (2014).
Lin, H.C. et al. AGORA: Assembly Guided by Optical Restriction Alignment. BMC Bioinformatics 13, 189 (2012).
Myers, E.W. The fragment assembly string graph. Bioinformatics 21 (suppl. 2), ii79–ii85 (2005).
Kuleshov, V. et al. Whole-genome haplotyping using long reads and statistical methods. Nat. Biotechnol. 32, 261–266 (2014).
Antonacci, F. et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 46, 1293–1302 (2014).
Gu, W., Zhang, F. & Lupski, J.R. Mechanisms for human genomic rearrangements. Pathogenetics 1, 4 (2008).
Sharp, A.J., Cheng, Z. & Eichler, E.E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 7, 407–442 (2006).
Bashir, A., Volik, S., Collins, C., Bafna, V. & Raphael, B.J. Evaluation of paired-end sequencing strategies for detection of genome rearrangements in cancer. PLoS Comput. Biol. 4, e1000051 (2008).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (2005).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, S. et al. SOAPindel: Efficient identification of indels from short paired reads. Genome Res. 23, 195–200 (2013).
Iskow, R.C. et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261 (2010).
Fuentes Fajardo, K.V. et al. Detecting false-positive signals in exome sequencing. Hum. Mutat. 33, 609–613 (2012).
Nguyen, J.V. Genomic Mapping: A Statistical and Algorithmic Analysis of the Optical Mapping System. PhD thesis, Univ. Southern California (2010).
Anantharaman, T. & Mishra, B. in Algorithms Bioinformatics WABI (eds. Gascuel, O. & Moret, B.M.E.) 27–40 (Springer, 2001).
Valouev, A., Schwartz, D.C., Zhou, S. & Waterman, M.S. An algorithm for assembly of ordered restriction maps from single DNA molecules. Proc. Natl. Acad. Sci. USA 103, 15770–15775 (2006).
Chaisson, M.J. & Tesler, G. Mapping single molecule sequencing reads using Basic Local Alignment with Successive Refinement (BLASR): theory and application. BMC Bioinformatics 13, 238 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at http://arxiv.org/abs/1207.3907 (2012).
English, A.C., Salerno, W.J. & Reid, J.G. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 15, 180 (2014).
Gotoh, O. An improved algorithm for matching biological sequences. J. Mol. Biol. 162, 705–708 (1982).
Eppstein, D., Galil, Z., Giancarlo, R. & Italiano, G.F. Sparse dynamic programming I: linear cost functions. J. ACM 39, 519–545 (1992).
Brudno, M. et al. Glocal alignment: finding rearrangements during alignment. Bioinformatics 19, i54–i62 (2003).
Dubchak, I., Poliakov, A., Kislyuk, A. & Brudno, M. Multiple whole-genome alignments without a reference organism. Genome Res. 19, 682–689 (2009).
Lee, C. Generating consensus sequences from partial order multiple sequence alignment graphs. Bioinformatics 19, 999–1008 (2003).
Wheeler, T.J. et al. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 41, D70–D82 (2013).
Bansal, V. & Bafna, V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics 24, i153–i159 (2008).
Carneiro, M.O. et al. Pacific Biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics 13, 375 (2012).
Koressaar, T. & Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291 (2007).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Acknowledgements
This work was supported in part by institutional support from the Icahn Institute for Genomics and Multiscale Biology, R01 HG005946, U01 HL107388, R01 DK098242-01, R01 MH106531, US National Institutes of Health (NIH) U41HG007497, the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts, the STARR Consortium, the WorldQuant Foundation, the Pershing Square Foundation, the Genomics & Epigenomics Core Facilities and SMRT Sequencing Center at Weill Cornell Medical College, and through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai. DNA samples were provided by the Coriell Institute for Medical Research and the US National Institute of Standards and Technology (NIST). We would also like to thank T. Zichner for assistance with the design of validations and M. Chaisson for assistance with running Blasr, the assembly-based SV pipeline, and in performing the CHM1 comparison.
Author information
Authors and Affiliations
Contributions
E.E.S., A.B., R.S., C.E.M., W.R.M. and R.B.D. conceived the project and provided resources for sequencing and algorithmic analysis. A.B. and E.E.S. provided bioinformatics oversight. J.O.K., M.H.-Y.F., A.M.S. and T.R. performed Illumina SV analysis and PCR validation. R.S., M.P. and E.E.P. prepared long libraries for PacBio sequencing. R.S., Y.G., A.C., S.C.B., R.A. and R.E.D. performed PacBio sequencing and primary analysis of hdf5 data. A.W.C.P., H.D., A.H., T.A., W.S., H.C. and P.-Y.K. generated the BioNano Data, built initial Genome Maps and performed BioNano alignment and SV calling. O.F., A.B. and M.P. performed PacBio SV analysis and validation. A.U., A.B. and C.-S.C. performed error correction and assembly. A.W.C.P. and H.D. built the initial hybrid scaffolding pipeline. A.B. and M.P. refined the hybrid scaffolding pipeline. A.W.C.P., H.D. and A.B. performed scaffold analysis and phasing. A.B., A.W.C.P., M.P. and A.U. generated figures for the main text. A.B., E.E.S., R.S., M.P. and A.W.C.P. primarily wrote the manuscript, though many coauthors provided edits and methods sections.
Corresponding author
Ethics declarations
Competing interests
A.W.C.P., H.C., W.S., T.A., A.H. and H.D. are employees of BioNano Genomics. C.-S.C., Y.G. and E.E.P. are employees of Pacific Biosciences, and E.E.S. is on the scientific advisory board of Pacific Biosciences.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1–4 and 6–12, Supplementary Results and Supplementary Notes 1–3 (PDF 16327 kb)
Supplementary Software
Custom scripts for performing hybrid scaffolding and SV analysis (ZIP 23658 kb)
Supplementary Table 5
Insertion and deletion SVs with phasing (XLSX 1079 kb)
Supplementary Table 13
Alignment coordinates between sequence contigs and V2 hybrid scaffolds (XLSX 260 kb)
Supplementary Table 14
Alignment coordinates between BioNano genome maps and V2 hybrid scaffolds (XLSX 89 kb)
Rights and permissions
About this article
Cite this article
Pendleton, M., Sebra, R., Pang, A. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods 12, 780–786 (2015). https://doi.org/10.1038/nmeth.3454
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3454
This article is cited by
-
The genomes of Vischeria oleaginous microalgae shed light on the molecular basis of hyper-accumulation of lipids
BMC Biology (2023)
-
SPUMONI 2: improved classification using a pangenome index of minimizer digests
Genome Biology (2023)
-
Pairwise comparative analysis of six haplotype assembly methods based on users’ experience
BMC Genomic Data (2023)
-
De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis
Nature Genetics (2023)
-
Long range PCR-based deep sequencing for haplotype determination in mixed HCMV infections
BMC Genomics (2022)