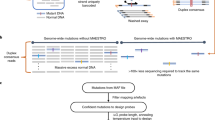
Abstract
The detection of minority variants in mixed samples requires methods for enrichment and accurate sequencing of small genomic intervals. We describe an efficient approach based on sequential rounds of hybridization with biotinylated oligonucleotides that enables more than 1-million-fold enrichment of genomic regions of interest. In conjunction with error-correcting double-stranded molecular tags, our approach enables the quantification of mutations in individual DNA molecules.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Schmitt, M.W., Prindle, M.J. & Loeb, L.A. Ann. NY Acad. Sci. 1267, 110–116 (2012).
Mamanova, L. et al. Nat. Methods 7, 111–118 (2010).
Hardenbol, P. et al. Nat. Biotechnol. 21, 673–678 (2003).
Kanagawa, T. J. Biosci. Bioeng. 96, 317–323 (2003).
Fox, E.J., Reid-Bayliss, K.S., Emond, M.J. & Loeb, L.A. Next Gener. Seq. Appl. 1, 106–109 (2014).
Glenn, T.C. Mol. Ecol. Resour. 11, 759–769 (2011).
Jabara, C.B., Jones, C.D., Roach, J., Anderson, J.A. & Swanstrom, R. Proc. Natl. Acad. Sci. USA 108, 20166–20171 (2011).
Kinde, I., Wu, J., Papadopoulos, N., Kinzler, K.W. & Vogelstein, B. Proc. Natl. Acad. Sci. USA 108, 9530–9535 (2011).
Hiatt, J.B., Pritchard, C.C., Salipante, S.J., O'Roak, B.J. & Shendure, J. Genome Res. 23, 843–854 (2013).
Schmitt, M.W. et al. Proc. Natl. Acad. Sci. USA 109, 14508–14513 (2012).
Soverini, S. et al. Leuk. Res. 38, 10–20 (2014).
Lou, D.I. et al. Proc. Natl. Acad. Sci. USA 110, 19872–19877 (2013).
Sweasy, J.B., Lauper, J.M. & Eckert, K.A. Radiat. Res. 166, 693–714 (2006).
Albertini, R.J., Nicklas, J.A., O'Neill, J.P. & Robison, S.H. Annu. Rev. Genet. 24, 305–326 (1990).
Kunkel, T.A. J. Biol. Chem. 279, 16895–16898 (2004).
Esposito, A. et al. Cancer Treat. Rev. 40, 648–655 (2014).
Buckley, S.A., Appelbaum, F.R. & Walter, R.B. Bone Marrow Transplant. 48, 630–641 (2013).
Alexandrov, L.B. et al. Nature 500, 415–421 (2013).
Kennedy, S.R., Salk, J.J., Schmitt, M.W. & Loeb, L.A. PLoS Genet. 9, e1003794 (2013).
Kennedy, S.R. et al. Nat. Protoc. 9, 2586–2606 (2014).
Burgess, D. et al. SeqCap EZ Library: Technical Note http://www.nimblegen.com/products/lit/06870406001_NG_SeqCapEZ_DoubleCaptureSR_20Aug2012.pdf (Roche NimbleGen, (2012)).
Robinson, J.T. et al. Nat. Biotechnol. 29, 24–26 (2011).
Egan, D.N., Beppu, L. & Radich, J.P. Biol. Blood Marrow Transplant. 21, 184–189 (2014).
Acknowledgements
Research reported in this publication was supported by the US National Institutes of Health under award numbers NCI P01-CA77852, R01-CA160674 and R33-CA181771 to L.A.L. and NCI U10-CA180861, P01-CA018029, R01-CA175008 and R01-CA175215 to J.P.R. We thank T. Walsh and M. Lee for assistance with DNA sequencing.
Author information
Authors and Affiliations
Contributions
M.W.S., E.J.F., M.J.P., K.S.R.-B., L.D.T., J.P.R. and L.A.L. contributed to experimental design. M.W.S., E.J.F. and M.J.P. performed the experiments in the paper and analyzed data. E.J.F., L.D.T. and J.P.R. contributed patient samples. M.W.S. and L.A.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.W.S. and L.A.L. are named on a patent application regarding duplex sequencing that has been filed by the University of Washington.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Tables 1–3 (PDF 266 kb)
Rights and permissions
About this article
Cite this article
Schmitt, M., Fox, E., Prindle, M. et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat Methods 12, 423–425 (2015). https://doi.org/10.1038/nmeth.3351
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3351
This article is cited by
-
Potential clinical utility of liquid biopsies in ovarian cancer
Molecular Cancer (2022)
-
Prognostic and therapeutic implications of measurable residual disease in acute myeloid leukemia
Journal of Hematology & Oncology (2021)
-
Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands
Nature Biotechnology (2021)
-
Commercial ctDNA Assays for Minimal Residual Disease Detection of Solid Tumors
Molecular Diagnosis & Therapy (2021)
-
Family reunion via error correction: an efficient analysis of duplex sequencing data
BMC Bioinformatics (2020)