Abstract
HISAT (hierarchical indexing for spliced alignment of transcripts) is a highly efficient system for aligning reads from RNA sequencing experiments. HISAT uses an indexing scheme based on the Burrows-Wheeler transform and the Ferragina-Manzini (FM) index, employing two types of indexes for alignment: a whole-genome FM index to anchor each alignment and numerous local FM indexes for very rapid extensions of these alignments. HISAT's hierarchical index for the human genome contains 48,000 local FM indexes, each representing a genomic region of ∼64,000 bp. Tests on real and simulated data sets showed that HISAT is the fastest system currently available, with equal or better accuracy than any other method. Despite its large number of indexes, HISAT requires only 4.3 gigabytes of memory. HISAT supports genomes of any size, including those larger than 4 billion bases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457, 1028–1032 (2009).
Cabili, M.N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Kim, D. & Salzberg, S.L. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 12, R72 (2011).
Garber, M., Grabherr, M.G., Guttman, M. & Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477 (2011).
Grant, G.R. et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 27, 2518–2528 (2011).
Engström, P.G. et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods 10, 1185–1191 (2013).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Wu, T.D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Burrows, M. & Wheeler, D.J.A. Block-sorting lossless data compression algorithm (Technical report 124). (Digital Equipment Corp., Palo Alto, 1994).
Ferragina, P. & Manzini, G. in Proc. 41st Annual Symp. Found. Comput. Sci. 390–398 (IEEE, 2000).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Wu, J., Anczuków, O., Krainer, A.R., Zhang, M.Q. & Zhang, C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res. 41, 5149–5163 (2013).
Griebel, T. et al. Modelling and simulating generic RNA-Seq experiments with the flux simulator. Nucleic Acids Res. 40, 10073–10083 (2012).
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012).
Acknowledgements
We thank G. Pertea and L. Song for their invaluable contributions to our discussions on HISAT. We also thank C. Trapnell for the use of his TuxSim simulation program. This work was supported in part by the National Human Genome Research Institute (US National Institutes of Health) under grants R01-HG006102 and R01-HG006677 to S.L.S.
Author information
Authors and Affiliations
Contributions
D.K., B.L. and S.L.S. performed the analysis and discussed the results of HISAT. D.K. implemented HISAT. D.K., B.L. and S.L.S. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Alignment speed and sensitivity of spliced alignment software for 40 million error-free simulated paired-end reads (100 bp long, 20 million pairs).
This figure shows the alignment speed and sensitivity for each type of pair (all, M, 2M_gt_15, 2M_8_15, 2M_1_7, gt_2M, where ‘all’ includes all the pair types). Since a pair consists of left and right reads, the type of a pair is determined by the more difficult read type. The difficulties of read types are given in the following order from easiest to most difficult: M, 2M_gt_15, 2M_8_15, 2M_1_7, and gt_2M. The plot on the left shows the alignment speed of the programs in terms of the number of pairs processed per second. The right plot shows alignment sensitivity. Pairs are categorized as: (1) correctly and uniquely mapped, (2) correctly mapped (multi-mapped), (3) incorrectly mapped, and (4) unmapped. Case (2) covers instances where an aligner mapped a pair to multiple locations and one of the locations was correct. These four categories encompass all of the pairs. The numbers in the right plot represent the percentages of case (1). The numbers inside the parentheses represent the percentages of cases (1) and (2) combined.
Supplementary Figure 2 Alignment speed and sensitivity of spliced alignment software for 20 million simulated single-end reads with a mismatch rate of 0.5% (100 bp long).
This figure shows the alignment speed and sensitivity for each type of read (all, M, 2M_gt_15, 2M_8_15, 2M_1_7, gt_2M, where ‘all’ includes all the read types). The plot on the left shows the alignment speed of the programs in terms of the number of reads processed per second. The right plot shows alignment sensitivity. Reads are categorized as: (1) correctly and uniquely mapped, (2) correctly mapped (multi-mapped), (3) incorrectly mapped, and (4) unmapped. Case (2) covers instances where an aligner mapped a read to multiple locations and one of the locations was correct. These four categories encompass all of the reads. The numbers in the right plot represent the percentages of case (1). The numbers inside the parentheses represent the percentages of cases (1) and (2) combined. Note that by looking at both plots, it is easy to see tradeoffs between alignment speed and sensitivity.
Supplementary Figure 3 Alignment results for 109 million reads, each 101 bp long, from a human sample.
Shown are the cumulative numbers of alignments up to a given edit distance. Edit distance is defined here simply as the number of differences (‘edits’) between the read and the reference sequence. The leftmost panel shows reads that matched exactly (with an edit distance of 0). The next panel (labelled “1”) shows the number of reads that aligned with either 0 or 1 mismatches; similarly for the panels labelled 2 and 3. Note that GSNAP and STAR report soft-clipped alignments where bases on the ends of reads are left unaligned. To compute edit distances for these alignments, we re-aligned the soft-clipped bases to their corresponding locations in the reference genome and calculated the number of mismatches.
Supplementary Figure 4 Alignment results of spliced alignment software for 109 million real reads (101 bp long).
This figure shows the cumulative number of spliced alignments up to a given edit distance (0, 1, 2, and 3) whose splice sites are known in gene annotations.
Supplementary Figure 5 Alignment speed and sensitivity of spliced-alignment software for 40 million simulated paired-end reads with a mismatch rate of 0.5% (100 bp long, 20 million pairs).
This figure shows the alignment speed and sensitivity for each type of pair (all, M, 2M_gt_15, 2M_8_15, 2M_1_7, gt_2M, where ‘all’ includes all the pair types). Since a pair consists of left and right reads, the type of a pair is determined by the more difficult read type. The difficulties of read types are given in the following order from easiest to most difficult: M, 2M_gt_15, 2M_8_15, 2M_1_7, and gt_2M. The plot on the left shows the alignment speed of the programs in terms of the number of pairs processed per second. The right plot shows alignment sensitivity. Pairs are categorized as: (1) correctly and uniquely mapped, (2) correctly mapped (multi-mapped), (3) incorrectly mapped, and (4) unmapped. Case (2) covers instances where an aligner mapped a pair to multiple locations and one of the locations was correct. These four categories encompass all of the pairs. The numbers in the right plot represent the percentages of case (1). The numbers inside the parentheses represent the percentages of cases (1) and (2) combined.
Supplementary Figure 6 Alignment results of spliced alignment software for ~218 million real paired-end reads (~109 million pairs).
This figure shows two plots: (1) the cumulative number of alignments up to a given edit distance (0, 1, 2, and 3) and (2) the cumulative number of spliced alignments whose splice sites are known in gene annotations. Note these alignments are pair alignments with the combined edit distance from the left and the right alignments. Spliced alignments are those whose read alignment is a spliced alignment.
Supplementary Figure 7 Alignment results of spliced alignment software for ~126 million real paired-end reads (~63 million pairs).
This figure shows two plots: (1) the cumulative number of alignments up to a given edit distance (0, 1, 2, and 3) and (2) the cumulative number of spliced alignments whose splice sites are known in gene annotations. Note these alignments are pair alignments with the combined edit distance from the left and the right alignments. Spliced alignments are those whose read alignment is a spliced alignment.
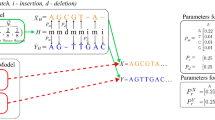
Supplementary Figure 8 Three working examples demonstrating how HISAT applies its hierarchical indexing for fast and sensitive alignment.
The examples include alignment of one exonic read and two junction reads (one an intermediate-anchored read and the other a long-anchored read). Reads are error-free and 100-bp long.
Supplementary Figure 9 Two-step approach version of HISAT to allow alignment of junction reads with small anchors.
This figure shows how to align reads with short anchors (1-7 bp) by making use of splice sites found by reads with long anchors.
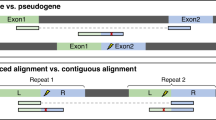
Supplementary Figure 10 Alignment of junction reads in the presence of processed pseudogenes.
This figure shows how to correctly align reads that would otherwise be mapped incorrectly to processed pseudogenes.
Supplementary Figure 11 Three more examples demonstrating how HISAT applies its hierarchical indexing for reads involving mismatches, indels and three exons.
The examples include alignment of one exonic read with one mismatch, one exonic read with an indel, and three exon spanning reads with two small anchors on both sides. Reads are 100-bp long.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–7 and Supplementary Note (PDF 453 kb)
Source data
Rights and permissions
About this article
Cite this article
Kim, D., Langmead, B. & Salzberg, S. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 (2015). https://doi.org/10.1038/nmeth.3317
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3317
This article is cited by
-
The fusion of multi-omics profile and multimodal EEG data contributes to the personalized diagnostic strategy for neurocognitive disorders
Microbiome (2024)
-
Genome-wide investigation of the TGF-β superfamily in scallops
BMC Genomics (2024)
-
Impacts of longitudinal water curtain cooling system on transcriptome-related immunity in ducks
BMC Genomics (2024)
-
Small-molecule agonist AdipoRon alleviates diabetic retinopathy through the AdipoR1/AMPK/EGR4 pathway
Journal of Translational Medicine (2024)
-
Integrated genome-wide association and transcriptomic analysis to identify receptor kinase genes to stripe rust resistance in wheat germplasm from southwestern China
BMC Plant Biology (2024)