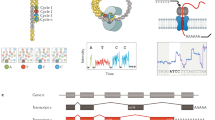
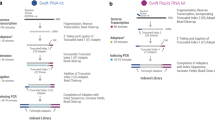
Abstract
Although RNA-seq is a powerful tool, the considerable time and cost associated with library construction has limited its utilization for various applications. RNAtag-Seq, an approach to generate multiple RNA-seq libraries in a single reaction, lowers time and cost per sample, and it produces data on prokaryotic and eukaryotic samples that are comparable to those generated by traditional strand-specific RNA-seq approaches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Wang, Z., Gerstein, M. & Snyder, M. Nat. Rev. Genet. 10, 57–63 (2009).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Nat. Methods 5, 621–628 (2008).
Wilhelm, B.T. et al. Nature 453, 1239–1243 (2008).
McHugh, C.A., Russell, P. & Guttman, M. Genome Biol. 15, 203 (2014).
Garber, M., Grabherr, M.G., Guttman, M. & Trapnell, C. Nat. Methods 8, 469–477 (2011).
Nagalakshmi, U. et al. Science 320, 1344–1349 (2008).
Levin, J.Z. et al. Nat. Methods 7, 709–715 (2010).
Garraway, L.A. & Lander, E.S. Cell 153, 17–37 (2013).
Golub, T.R. et al. Science 286, 531–537 (1999).
Amit, I. et al. Science 326, 257–263 (2009).
Lamb, J. et al. Science 313, 1929–1935 (2006).
Ravasi, T. et al. Cell 140, 744–752 (2010).
Hashimshony, T., Wagner, F., Sher, N. & Yanai, I. Cell Rep. 2, 666–673 (2012).
Jaitin, D.A. et al. Science 343, 776–779 (2014).
Islam, S. et al. Genome Res. 21, 1160–1167 (2011).
Kivioja, T. et al. Nat. Methods 9, 72–74 (2012).
Islam, S. et al. Nat. Methods 11, 163–166 (2014).
Anders, S. & Huber, W. Genome Biol. 11, R106 (2010).
Barczak, A.K. et al. Proc. Natl. Acad. Sci. USA 109, 6217–6222 (2012).
Michel, B. PLoS Biol. 3, e255 (2005).
Giannoukos, G. et al. Genome Biol. 13, R23 (2012).
Adiconis, X. et al. Nat. Methods 10, 623–629 (2013).
Guttman, M., Russell, P., Ingolia, N.T., Weissman, J.S. & Lander, E.S. Cell 154, 240–251 (2013).
Haas, B.J., Chin, M., Nusbaum, C., Birren, B.W. & Livny, J. BMC Genomics 13, 734 (2012).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
Gardner, P.P. et al. Nucleic Acids Res. 37, D136–D140 (2009).
Novichkov, P.S. et al. BMC Genomics 14, 745 (2013).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Genome Biol. 10, R25 (2009).
Barnett, D.W., Garrison, E.K., Quinlan, A.R., Stromberg, M.P. & Marth, G.T. Bioinformatics 27, 1691–1692 (2011).
Li, B. & Dewey, C.N. BMC Bioinformatics 12, 323 (2011).
Langmead, B. & Salzberg, S.L. Nat. Methods 9, 357–359 (2012).
Busby, M.A., Stewart, C., Miller, C.A., Grzeda, K.R. & Marth, G.T. Bioinformatics 29, 656–657 (2013).
Love, M.I., Huber, W. & Anders, S. Genome Biol. 15, 550 (2014).
Jiao, X. et al. Bioinformatics 28, 1805–1806 (2012).
Acknowledgements
We thank all members of Lander and Guttman labs for their help, especially J.M. Engreitz, P. McDonel and K. Sirokman; L. Gaffney for assistance with figures; and C. Nusbaum for helpful suggestions on the manuscript. We thank I. Antoshechkin and the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech. This work was supported by a US National Institutes of Health (NIH) Director's Early Independence Award (DP5OD012190 to M. Guttman), funds from the Broad Institute of MIT and Harvard and the California Institute of Technology (M. Guttman) and funds from the US National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services, under contract no. HHSN272200900018C.
Author information
Authors and Affiliations
Contributions
A.A.S. and M. Guttman conceived of the approach and developed the initial RNAtag-Seq protocol; A.A.S., C.S., G.G., D.C. and A.G. optimized and streamlined the RNAtag-Seq protocol; A.A.S., G.G., D.C., R.P.B., R.F.R. and M.M.P. generated RNA-seq libraries; A.K., J.C., N.N., M.B., M. Garber, M. Guttman and J.L. analyzed data; R.P.B., D.T.H., M. Guttman and J.L. designed differential expression experiments; M. Guttman and J.L. supervised the project and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
A.A.S., C.S. and M. Guttman are inventors on a provisional patent owned by the Broad Institute covering massive-scale barcoding and sequencing of RNA samples.
Integrated supplementary information
Supplementary Figure 1 Comparison of replicate data sets generated by dUTP and RNAtag-Seq for K562 and three bacterial species.
(a) Scatter plots of log10 of normalized abundance (TPM for K562 and FPKMO (fragments per kilobase of ORF per million fragments aligned to all ORFs) for bacteria). (b) Distribution of reads aligning across the lengths of transcripts or ORFs. (c) Percent of all genes detected per aligned read pairs.
Supplementary Figure 2 Read coverage in RNAtag-Seq and dUTP data sets over the same genomic region of E. coli.
Values of coverage per genomic position are shown in arbitrary units. Coverage on the plus and minus strand are shown in blue and red, respectively.
Supplementary Figure 3 Distribution of reads per barcode in a single pool.
(a) Results of individual ligation with set of 32 barcodes reproducibly yielding total reads within 3-fold. These pools were sequenced on the Illumina HiSeq platform. Dotted lines correspond to number of reads expected with even distribution across all barcodes. (b) Total reads among 54 individually barcoded RNAtag-Seq libraries in a single pool. This pool was sequenced on the Illumina MiSeq platform. Dotted lines correspond number of reads expected with even distribution across all barcodes.
Supplementary Figure 5 Comparison of RNAtag-Seq data generated with 54 RNA and 32 DNA adaptors.
(a) Frequency of all possible dinucleotide sequences following adaptor sequences in paired end reads and in all annotated E. coli ORFs. (b) Correlation plots of normalized abundance per gene from replicate RNA samples tagged with RNA and DNA adaptors
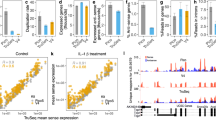
Supplementary Figure 6 Additional results from differential gene expression analysis of E. coli using RNAtag-Seq.
(a) MA plots of ciprofloxacin susceptible (CipS, left panel) and resistant (CipR, right panel) E. coli clinical isolates 10 and 60 minutes after exposure to ciprofloxacin vs. untreated. Genes significantly up- and down-regulated (> 3 fold, Padj < 0.05) by are highlighted in black. Genes in the SOS regulon are labeled in red. (b) Venn diagrams of genes significantly differentially expressed (> 3 fold, Padj < 0.05) after exposure of CipS to ciprofloxacin or between early and late growth phases in untreated CipS cultures.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Protocol (PDF 1356 kb)
Supplementary Table 1
Metrics for RNAtagSeq and dUTP data (XLSX 57 kb)
Supplementary Table 2
Validated sets of RNAtag-Seq adapters used in this study (XLSX 40 kb)
Supplementary Table 3
FPKMO correlations (R2) of replicate libraries barcoded with different RNA adaptors (XLSX 49 kb)
Supplementary Table 4
Normalized expression values (TPM) for 11 mouse tissues (XLSX 3497 kb)
Supplementary Table 5
Lists of genes differentially expressed in mouse tissues (XLSX 59 kb)
Supplementary Table 6
Normalized expression values (fragments per kb of gene per million of reads mapped to ORFs) for 32 E. coli clinical isolates in culture with and without ciprofloxacin (XLSX 1579 kb)
Supplementary Table 7
Lists of genes up- and downregulated in cipro susceptible strains following exposure to cipro for the indicated times (XLSX 36 kb)
Rights and permissions
About this article
Cite this article
Shishkin, A., Giannoukos, G., Kucukural, A. et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12, 323–325 (2015). https://doi.org/10.1038/nmeth.3313
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3313
This article is cited by
-
High-throughput transcriptomics of 409 bacteria–drug pairs reveals drivers of gut microbiota perturbation
Nature Microbiology (2024)
-
Systemic metabolic engineering of Enterobacter aerogenes for efficient 2,3-butanediol production
Applied Microbiology and Biotechnology (2024)
-
First report of Citrus concave gum-associated virus (CCGaV) on apple (Malus spp.) in South Africa
Journal of Plant Pathology (2024)
-
RNA-seq research landscape in Africa: systematic review reveals disparities and opportunities
European Journal of Medical Research (2023)
-
Temporal-iCLIP captures co-transcriptional RNA-protein interactions
Nature Communications (2023)