Abstract
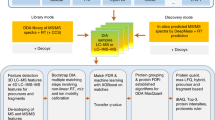
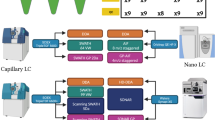
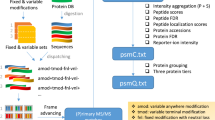
As a result of recent improvements in mass spectrometry (MS), there is increased interest in data-independent acquisition (DIA) strategies in which all peptides are systematically fragmented using wide mass-isolation windows ('multiplex fragmentation'). DIA-Umpire (http://diaumpire.sourceforge.net/), a comprehensive computational workflow and open-source software for DIA data, detects precursor and fragment chromatographic features and assembles them into pseudo–tandem MS spectra. These spectra can be identified with conventional database-searching and protein-inference tools, allowing sensitive, untargeted analysis of DIA data without the need for a spectral library. Quantification is done with both precursor- and fragment-ion intensities. Furthermore, DIA-Umpire enables targeted extraction of quantitative information based on peptides initially identified in only a subset of the samples, resulting in more consistent quantification across multiple samples. We demonstrated the performance of the method with control samples of varying complexity and publicly available glycoproteomics and affinity purification–MS data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bantscheff, M., Lemeer, S., Savitski, M.M. & Kuster, B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 404, 939–965 (2012).
Nesvizhskii, A.I. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics 73, 2092–2123 (2010).
Bailey, D.J., McDevitt, M.T., Westphall, M.S., Pagliarini, D.J. & Coon, J.J. Intelligent data acquisition blends targeted and discovery methods. J. Proteome Res. 13, 2152–2161 (2014).
Weisbrod, C.R., Eng, J.K., Hoopmann, M.R., Baker, T. & Bruce, J.E. Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. J. Proteome Res. 11, 1621–1632 (2012).
Michalski, A., Cox, J. & Mann, M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 (2011).
Gillet, L.C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012).
Tate, S., Larsen, B., Bonner, R. & Gingras, A.C. Label-free quantitative proteomics trends for protein-protein interactions. J. Proteomics 81, 91–101 (2013).
Venable, J.D., Dong, M.Q., Wohlschlegel, J., Dillin, A. & Yates, J.R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 (2004).
Silva, J.C., Gorenstein, M.V., Li, G.Z., Vissers, J.P. & Geromanos, S.J. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 (2006).
Panchaud, A. et al. Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 (2009).
Geiger, T., Cox, J. & Mann, M. Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics 9, 2252–2261 (2010).
Egertson, J.D. et al. Multiplexed MS/MS for improved data-independent acquisition. Nat. Methods 10, 744–746 (2013).
Distler, U. et al. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 11, 167–170 (2014).
Purvine, S., Eppel, J.T., Yi, E.C. & Goodlett, D.R. Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics 3, 847–850 (2003).
Colangelo, C.M., Chung, L., Bruce, C. & Cheung, K.H. Review of software tools for design and analysis of large scale MRM proteomic datasets. Methods 61, 287–298 (2013).
Röst, H.L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).
Rosenberger, G. et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci. Data 1, 140031 (2014).
Li, G.Z. et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 (2009).
Pak, H. et al. Clustering and filtering tandem mass spectra acquired in data-independent mode. J. Am. Soc. Mass Spectrom. 24, 1862–1871 (2013).
Craig, R., Cortens, J.P. & Beavis, R.C. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 (2004).
Eng, J.K., Jahan, T.A. & Hoopmann, M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013).
Kim, S. et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics 9, 2840–2852 (2010).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Shteynberg, D. et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690 (2011).
Nesvizhskii, A.I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Lambert, J.P. et al. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 (2013).
Lam, H. et al. Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 (2008).
Reiter, L. et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 (2011).
Liu, Y. et al. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol. Cell. Proteomics 13, 1753–1768 (2014).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Ludwig, C., Claassen, M., Schmidt, A. & Aebersold, R. Estimation of absolute protein quantities of unlabeled samples by selected reaction monitoring mass spectrometry. Mol. Cell. Proteomics 11, M111.013987 (2012).
Collins, B.C. et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods 10, 1246–1253 (2013).
Nesvizhskii, A.I. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics 12, 1639–1655 (2012).
Choi, H. et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 (2011).
Choi, H., Glatter, T., Gstaiger, M. & Nesvizhskii, A.I. SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J. Proteome Res. 11, 2619–2624 (2012).
Chatr-Aryamontri, A. et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 41, D816–D823 (2013).
Jeronimo, C. et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27, 262–274 (2007).
Prakash, A. et al. Hybrid data acquisition and processing strategies with increased throughput and selectivity: pSMART analysis for global qualitative and quantitative analysis. J. Proteome Res. 13, 5415–5430 (2014).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Chambers, M.C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Tautenhahn, R., Bottcher, C. & Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9, 504 (2008).
Nesvizhskii, A.I. et al. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol. Cell. Proteomics 5, 652–670 (2006).
Kryuchkov, F., Verano-Braga, T., Hansen, T.A., Sprenger, R.R. & Kjeldsen, F. Deconvolution of mixture spectra and increased throughput of peptide identification by utilization of intensified complementary ions formed in tandem mass spectrometry. J. Proteome Res. 12, 3362–3371 (2013).
Deutsch, E.W. et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 (2010).
Tsou, C.C. et al. IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol. Cell. Proteomics 9, 131–144 (2010).
Lam, H., Deutsch, E.W. & Aebersold, R. Artificial decoy spectral libraries for false discovery rate estimation in spectral library searching in proteomics. J. Proteome Res. 9, 605–610 (2010).
Cox, J., Michalski, A. & Mann, M. Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom. 22, 1373–1380 (2011).
Barsnes, H. et al. compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinformatics 12, 70 (2011).
Escher, C. et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121 (2012).
Vizcaíno, J.A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014).
Acknowledgements
We thank B. MacLean for help with Skyline, H. Röst and B. Collins for help with OpenSWATH and S. Tate for useful discussions. We also thank S. Danielson at Thermo Scientific for access to the Q Exactive Plus and Z.-Y. Lin for the acquisition of the DIA samples for MEPCE, EIF4A2 and GFP. This work was supported by US National Institutes of Health grants 5R01GM94231 (to A.I.N. and A.-C.G.), R01GM107148 and U24DK097153 (to A.I.N.); the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-069 to A.-C.G., A.I.N. and H.C.) and the Canadian Institutes of Health Research (MOP-84314 and MOP-123322 to A.-C.G.); and Singapore Ministry of Education grant R-608-000-088-112 (to H.C.).
Author information
Authors and Affiliations
Contributions
A.I.N. and C.-C.T. conceived the project and developed the algorithm. C.-C.T. implemented the software. D.A. assisted with the OpenSWATH analysis and contributed to the algorithm and software development. B.L. and M.T. acquired mass spectrometry data. H.C. assisted with SAINT scoring and contributed to the development of protein quantification strategies. A.-C.G., C.-C.T., B.L. and A.I.N. designed experiments and analyzed data. A.I.N. and A.-C.G. supervised the project. C.-C.T., A.I.N. and A.-C.G. wrote the manuscript with input from B.L. and D.A.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–29 (PDF 7641 kb)
Supplementary Table 1
Number of protein and peptide ion identifications from DDA MS/MS and DIA pseudo-MS/MS spectra obtained with three different search engines (X!Tandem, Comet and MSGF+), as well as with all three search engines combined (XLSX 12 kb)
Supplementary Table 2
Protein identification and quantification results (protein, peptide ion and fragment ion levels), human cell lysate samples (XLSX 30210 kb)
Supplementary Table 3
Protein identification and quantification results (protein, peptide ion and fragment ion levels), E. coli cell lysate samples (XLSX 20544 kb)
Supplementary Table 4
Protein identification and quantification results (protein, peptide ion and fragment ion levels), UPS2 samples (XLSX 3843 kb)
Supplementary Table 5
Peptide ion identifications from DDA, DIA with DIA-Umpire and DIA with OpenSWATH, human cell lysate samples (XLSX 2051 kb)
Supplementary Table 6
Peptide ion identifications from DDA, DIA with DIA-Umpire and DIA with OpenSWATH, E. coli cell lysate samples (XLSX 1394 kb)
Supplementary Table 7
Protein identification and quantification results (protein, peptide ion and fragment ion levels), DIA (SWATH) glycoproteomics data set (XLSX 19282 kb)
Supplementary Table 8
Protein identification and quantification results (protein, peptide ion and fragment ion levels), AP-SWATH interactome data set (XLSX 42939 kb)
Supplementary Table 9
SAINT results, AP-SWATH interactome data set (XLSX 119 kb)
Supplementary Table 10
Number of peptide ions and proteins for a representative DIA (SWATH) run identified using different thresholds for precursor-fragment grouping (XLSX 12 kb)
Supplementary Table 11
List of the raw mass spectrometry files (XLSX 12 kb)
Rights and permissions
About this article
Cite this article
Tsou, CC., Avtonomov, D., Larsen, B. et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods 12, 258–264 (2015). https://doi.org/10.1038/nmeth.3255
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3255