Abstract
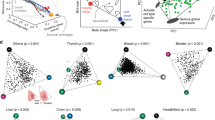
We present the Pareto task inference method (ParTI; http://www.weizmann.ac.il/mcb/UriAlon/download/ParTI) for inferring biological tasks from high-dimensional biological data. Data are described as a polytope, and features maximally enriched closest to the vertices (or archetypes) allow identification of the tasks the vertices represent. We demonstrate that human breast tumors and mouse tissues are well described by tetrahedrons in gene expression space, with specific tumor types and biological functions enriched at each of the vertices, suggesting four key tasks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kim, H.D., Shay, T., O'Shea, E.K. & Regev, A. Science 325, 429–432 (2009).
Kalisky, T., Blainey, P. & Quake, S.R. Annu. Rev. Genet. 45, 431–445 (2011).
Curtis, C. et al. Nature 486, 346–352 (2012).
Bendall, S.C. & Nolan, G.P. Nat. Biotechnol. 30, 639–647 (2012).
The Cancer Genome Atlas Network. Nature 490, 61–70 (2012).
Ringnér, M. Nat. Biotechnol. 26, 303–304 (2008).
Van der Maaten, L. & Hinton, G. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Hastie, T., Tibshirani, R. & Friedman, J. in The Elements of Statistical Learning 2nd edn. 520–528 (Springer, 2009).
Shoval, O. et al. Science 336, 1157–1160 (2012).
Sheftel, H., Shoval, O., Mayo, A. & Alon, U. Ecol. Evol. 3, 1471–1483 (2013).
Szekely, P., Sheftel, H., Mayo, A. & Alon, U. PLoS Comput. Biol. 9, e1003163 (2013).
Mørup, M. & Hansen, L.K. Neurocomputing 80, 54–63 (2012).
Li, J. & Bioucas-Dias, J.M. IEEE Int. Geosci. Remote Sens. Symp. 3, 250–253 (2008).
Chan, T.-H., Chi, C.-Y., Huang, Y.-M. & Ma, W.-K. IEEE Trans. Signal Process. 57, 4418–4432 (2009).
Chan, T.-H., Liou, J.-Y., Ambikapathi, A., Ma, W.-K. & Chi, C.-Y. in Proc. IEEE Int. Conf. Acoust. Speech Signal Process. 1237–1240 (IEEE, 2012).
Bioucas-Dias, J.M. et al. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 5, 354–379 (2012).
Schwartz, R. & Shackney, S.E. BMC Bioinformatics 11, 42 (2010).
Tolliver, D., Tsourakakis, C., Subramanian, A., Shackney, S. & Schwartz, R. Bioinformatics 26, i106–i114 (2010).
Thøgersen, J.C., Mørup, M., Damkiær, S., Molin, S. & Jelsbak, L. BMC Bioinformatics 14, 279 (2013).
Subramanian, A. et al. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Lehmann, B.D. et al. J. Clin. Invest. 121, 2750–2767 (2011).
Lattin, J.E. et al. Immunome Res. 4, 5 (2008).
Cutler, A. & Breiman, L. Technometrics 36, 338–347 (1994).
Bioucas-Dias, J.M. in Hyperspectral Image Signal Process. Evol. Remote Sens. First Workshop 1–4 (IEEE, 2009).
Mann, H.B. & Whitney, D.R. Ann. Math. Stat. 18, 50–60 (1947).
Benjamini, Y. & Hochberg, Y. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Nishimura, D. Biotech Softw. Internet Rep. 2, 117–120 (2001).
Kanehisa, M. & Goto, S. Nucleic Acids Res. 28, 27–30 (2000).
Croft, D. et al. Nucleic Acids Res. 39 (suppl. 1), D691–D697 (2011).
Acknowledgements
We thank N. Drayman, B. Towbin, M. Botzman, Y. Liron, M. Adler, G. Aidelberg, D. Rothschild, S. Malihi, O. Szekely and members of the Alon lab for discussions. We acknowledge support by the Human Frontier Science Program, project number RGP0020/2012, European Research Council, project number 249919, and Rising Tide Cancer Research Fund, project number 721176. U.A. receives support as the Abisch-Frenkel Professorial Chair. J.H. acknowledges the support of the Swiss National Science Foundation (PBBSP3_14961) and EMBO (ALTF 1160-2012).
Author information
Authors and Affiliations
Contributions
Y.H., H.S., J.H. and P.S. developed the method and analyzed the data. N.B.B.-M. analyzed the microarray breast cancer data. Y.K., A.T. and A.E.M. consulted on the method and algorithm. U.A. designed the method and research program. Y.H., H.S., J.H. and P.S. wrote the Matlab code, and Y.H., H.S., J.H., P.S. and U.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 The best trade-off phenotypes lie on polytopes in trait space.
(A) 2 tasks result in a line. (B) 3 tasks result in a triangle. (C) 4 tasks result in a tetrahedron.
Supplementary Figure 2 Schematic description of Pareto archetype analysis and its relation to clustering analysis.
(A) Clustering works well for data that is divided into discrete groups. (B) Data that uniformly fill a triangle is clustered by k-means clustering into three clusters, so that each data point is categorized into one of three categories. Close-by points (circled in black) can be assigned to different categories. (C) Archetype analysis of the same data provides a continuous description where each data point is described by the distances from the archetypes. Thus two near-by points (circled in black) are categorized in different clusters according to clustering algorithms, but have similar weights in ParTI. (D) Point density in the dataset affects clustering but not the archetypes of the ParTI method. Shown are two datasets, where clustering yields different clusters whereas the archetype positions remain unchanged.
Supplementary Figure 3 The breast cancer gene expression data set is well enclosed by a tetrahedron.
(A) Fraction of the total variance explained by the polytope as a function of the number of archetypes. Archetypes in dimension d (=#archetypes-1) were calculated using the PCHA algorithm. Explained variance was computed. The effective number of archetypes can be estimated from the maximal distance between the EV and the line connecting between the first and last points. (B) 3D plot of the data and enclosing tetrahedron. The axes are the first three principal components, which explain 30.4% of variance. The colored ellipsoids represent the archetype location and error on the most varying directions. Archetype error bars are obtained by bootstrapping. Each ellipsoid represents 68% confidence level. The inset near each archetype shows the projection of the data on the plane defined by the tetrahedron’s face opposing that archetype.
Supplementary Figure 4 The clusters defined by Curtis et al.3 are located in specific areas of the tetrahedron.
Each panel represents the location of one of the clusters found by Curtis et al. in the tetrahedron calculated by the ParTI method for the breast cancer dataset. The volume colored in purple is defined by the region in the polytope in which a given cluster is most enriched (the convex hull of the 50% locally enriched points). Data points belonging to the relevant cluster are plotted in black.
Supplementary Figure 5 Several features found by Enrichment At Archetype (EAA) are not maximal at the archetype.
Each panel shows the density of a given feature as a function of distance from the archetype to the features detected by using the archetype position (Enrichment At Archetype, EAA) instead of PartTI. As can be seen (red boxes), some features are maximally enriched at some distance from the archetype rather than at the archetype itself, suggesting that they are not associated with the archetype’s biological task according to Pareto optimality theory. x-axis is the bin number, y-axis is normalized enrichment (compared to the mean density).
Supplementary Figure 6 The “basal” archetype splits as additional archetypes are added to the analysis.
The tree of archetypes is determined by Euclidean distance between archetypes in different dimensions.
Supplementary Figure 7 Breast cancer gene expression data profiled by mRNA-seq is well described by a tetrahedron.
The axes are the first three principal components. The color ellipsoids represent the archetype location and error on the most varying directions. Archetype error bars are obtained by bootstrapping. Each ellipsoid represents 68% confidence level.
Supplementary Figure 8 Three-dimensional plot of the tissue data set and enclosing tetrahedron.
Axes are the first three principal components. Different tissue categories are marked by different coloring – neural (green), macrophage and microglia (purple), secretory glands (red), stem cells (light blue), hematopoietic cells (lymphoid – orange, myeloid – olive green), other homogeneous tissues (blue).
Supplementary Figure 9 Caveats in Pareto archetype analysis.
(A) Coin toss data (binomial process B(0.5,N)) falls on a triangular shaped region in log-log plots of number of heads versus number of tosses because variance increases with number of tosses. The triangle is unrelated to Pareto theory. No data point is expected to be enriched for any feature. (B) A non-convex distribution of data can result in significant triangle assignment. Pareto origin of this triangle can be doubted if no feature is enriched near archetype z. (C) A distant outlier from the rest of the dataset can make a triangle in the PCA, since first principal component will span the line between the outlier and the data and second component will condense the rest of the data into a line, thus forming an artificial triangle.
Supplementary Figure 10 The ‘elbow’ method.
We assess the best-fit number of archetypes automatically by plotting the explained variance (EV) vs. the number of archetypes: we look for the ‘elbow on the plot by seeking for the point farthest from the line connecting the first and last EV. Here we show an example for the mouse tissue dataset. This method should be tested for different maximal numbers of archetypes to assess its robustness.
Supplementary Figure 11 Different algorithms result in similar positions of the archetypes.
Breast cancer data plotted in the three first PCs space, each tissue sample is a black dot. Blue, green, red and yellow circles represent archetypes positions found by Sisal, MVSA, MVES and SDVMM, respectively.
Supplementary Figure 12 The position of the archetypes is robust to data sampling (cancer data set).
Shown are the positions of the archetypes found for the bootstrapped datasets using sampling with replacement (blue points) and the archetypes position when removing points from the convex hull of the data (red points).
Supplementary Figure 13 Three-dimensional plot of the data and enclosing tetrahedron of the mouse tissue data set.
The axes are the first three principal components that explain 61% of variance. The color ellipsoids represent the archetype location and error on the most varying directions. Archetype error bars are obtained by bootstrapping. Each ellipsoid represents 68% confidence level.
Supplementary Figure 14 Explained variance measures both dimensionality and structure of the data set.
The fraction of explained variance curves of a tetrahedron (red), a 3D-cube (green) and a 3D- sphere (blue) with 3% noise embedded in a 100 dimensional space. The explained variance was calculated with the PCHA algorithm for 2-10 archetypes.
Supplementary Figure 15 Archetypes in the real data set show many more enriched features than are expected by chance.
Purple circles indicate the number of enriched features at the most-enriched archetype in the shuffled dataset as a function of p-value threshold Pth, averaged over 1000 shuffled datasets. Black circles indicate the mean number of enriched features at a single archetype in the shuffled dataset, averaged over 1000 shuffled datasets. Brown, red, green and blue small circles indicate the corresponding total number of enriched features at the archetypes in the real non-shuffled dataset.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Table 6, Supplementary Notes 1–12, Supplementary Results and Supplementary Discussion (PDF 2111 kb)
Supplementary Table 1
List of clinical features in the breast cancer dataset. (XLS 33 kb)
Supplementary Table 2
Breast cancer enrichment analysis (microarrays). (XLS 103 kb)
Supplementary Table 3
Breast cancer enrichment with clustering of Curtis et al. (XLS 234 kb)
Supplementary Table 4
Breast cancer enrichment with Gaussian Mixture Model. (XLS 112 kb)
Supplementary Table 5
Breast cancer enrichment with K Means. (XLS 44 kb)
Supplementary Table 7
Archetype profiling using the method of Thøgersen et al. (XLS 114 kb)
Supplementary Table 8
Breast cancer enrichment analysis (RNAseq). (XLS 114 kb)
Supplementary Table 9
Mouse Tissues Enrichment Analysis (XLS 86 kb)
Source data
Rights and permissions
About this article
Cite this article
Hart, Y., Sheftel, H., Hausser, J. et al. Inferring biological tasks using Pareto analysis of high-dimensional data. Nat Methods 12, 233–235 (2015). https://doi.org/10.1038/nmeth.3254
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3254
This article is cited by
-
Network traits predict ecological strategies in fungi
ISME Communications (2022)
-
Proteomic traits vary across taxa in a coastal Antarctic phytoplankton bloom
The ISME Journal (2022)
-
Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology
Nature Cancer (2022)
-
Comparative assessment and novel strategy on methods for imputing proteomics data
Scientific Reports (2022)
-
Chronic nicotine increases midbrain dopamine neuron activity and biases individual strategies towards reduced exploration in mice
Nature Communications (2021)