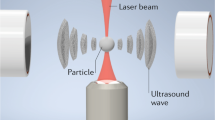
Abstract
Force spectroscopy has become an indispensable tool to unravel the structural and mechanochemical properties of biomolecules. Here we extend the force spectroscopy toolbox with an acoustic manipulation device that can exert forces from subpiconewtons to hundreds of piconewtons on thousands of biomolecules in parallel, with submillisecond response time and inherent stability. This method can be readily integrated in lab-on-a-chip devices, allowing for cost-effective and massively parallel applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bustamante, C., Cheng, W. & Meija, Y.X. Cell 144, 480–497 (2011).
Neuman, K.C. & Nagy, A. Nat. Methods 5, 491–505 (2008).
Herbert, K.M., Greenleaf, W.J. & Block, S.M. Annu. Rev. Biochem. 77, 149–176 (2008).
Zhao, Y. et al. Lab Chip 13, 2183–2198 (2013).
De Vlaminck, I. & Dekker, C. Annu. Rev. Biophys. 41, 453–472 (2012).
Kim, S., Blainey, P.C., Schroeder, C.M. & Xie, X.S. Nat. Methods 4, 397–399 (2007).
Halvorsen, K. & Wong, W.P. Biophys. J. 98, L53–L55 (2010).
Fazio, T., Visnapuu, M.-L., Wind, S. & Greene, E.C. Langmuir 24, 10524–10531 (2008).
Smith, S.B., Cui, Y. & Bustamante, C. Science 271, 795–799 (1996).
van Mameren, J. et al. Proc. Natl. Acad. Sci. USA 106, 18231–18236 (2009).
Settnes, M. & Bruus, H. Phys. Rev. E 85, 016327 (2012).
Heller, I. et al. Nat. Methods 10, 910–916 (2013).
Glynne-Jones, P., Boltryk, R.J. & Hill, M. Lab Chip 12, 1417–1426 (2012).
Laurens, N. et al. Nucleic Acids Res. 37, 5454–5464 (2009).
Odijk, T. Macromolecules 28, 7016–7018 (1995).
Chen, Z., Yang, H. & Pavletich, N.P. Nature 453, 489–494 (2008).
Fu, H., Le, S., Muniyappa, K. & Yan, J. PLoS ONE 8, e66712 (2013).
Merkel, R., Nassoy, P., Leung, A., Ritchie, K. & Evans, E. Nature 397, 50–53 (1999).
Neuert, G., Albrecht, C., Pamir, E. & Gaub, H.E. FEBS Lett. 580, 505–509 (2006).
Moayed, F., Mashaghi, A. & Tans, S.J. PLoS ONE 8, e54440 (2013).
Hill, M., Shen, Y. & Hawkes, J.J. Ultrasonics 40, 385–392 (2002).
Hill, M., Townsend, R.J. & Harris, N.R. Ultrasonics 48, 521–528 (2008).
Gosse, C. & Croquette, V. Biophys. J. 82, 3314–3329 (2002).
te Velthuis, A.J.W., Kerssemakers, J.W.J., Lipfert, J. & Dekker, N.H. Biophys. J. 99, 1292–1302 (2010).
Berg-Sørensen, K. & Flyvbjerg, H. Rev. Sci. Instrum. 75, 594–612 (2004).
Nørrelykke, S.F. & Flyvbjerg, H. Rev. Sci. Instrum. 81, 075103 (2010).
Han, L. et al. in Mathematics of DNA Structure. Function and Interactions (eds. Benham, C.J., Harvey, S., Olson, W.K., Sumners, D.W. & Swigon, D.) 123–138 (Springer, 2009).
Dunlap, D., Yokoyama, R. & Ling, H. Nucleic Acids Res. 40, 11281–11291 (2012).
Acknowledgements
We thank A.S. Biebricher, T.P. Hoekstra and S. d'Haene for biochemical help; R.J. Limburg and P.J.M. Caspers for technical support; N. Laurens for his work on the tracking software; O.D. Broekmans for his help with making the software available online; M. Hill and P. Glynne-Jones (University of Southampton) for providing the software comprising the one-dimensional acoustical model; and D.D. Dunlap (Emory University) for supplying the torsionally constrained DNA construct. This work is part of the research program of the Foundation for Fundamental Research on Matter (FOM) (E.J.G.P. and G.J.L.W.), which is part of the Netherlands Organization for Scientific Research (NWO). We acknowledge support by NWO VICI (E.J.G.P. and G.J.L.W.) as well as a European Research Council (ERC) starting grant (G.J.L.W.) and an ERC advanced grant (M.R.-M.).
Author information
Authors and Affiliations
Contributions
G.S., G.J.L.W. and E.J.G.P. initiated the development of acoustic force spectroscopy as a novel single-molecule technique rooted in discussions during a visit to the laboratory of G.T. and M.R.-M. G.T. and M.R.-M. provided the background knowledge and know-how on acoustic forces. G.S. conceived of and designed the instrument and research. G.S. and D.K. built the instrument, wrote the software, performed the experiments and analyzed the data. G.T., E.J.G.P. and G.J.L.W. advised on instrument design. E.J.G.P. and G.J.L.W. supervised the project. G.S., D.K., G.T., M.R.-M, E.J.G.P. and G.J.L.W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Accuracy of microsphere tracking in x, y and z.
Accuracy of tracking at 50 Hz in x, y, and z of a 4.5 µm diameter polystyrene microsphere (a) and a 1.5 µm diameter silica microsphere (b). These microspheres were stuck nonspecifically to the flow cell surface. To correct for mechanical drifts a reference microsphere (1.5 µm diameter silica) was tracked simultaneously and its position was subtracted. The standard deviations of the position distributions for the 4.5 µm diameter polystyrene microsphere and 1.5 µm diameter silica microsphere are σx,4.5 = 3 nm, σy,4.5 = 3 nm, σz,4.5 = 4 nm and σx1.5 = 2 nm, σy,1.5 = 1 nm, σz,1.5 = 5 nm respectively.
Supplementary Figure 2 The AFS device.
The piezo element, with wires connected to the electrodes, is glued onto the aluminum layer. A US quarter dollar coin is placed at the side for size perspective.
Supplementary Figure 3 Histogram of the force distribution.
The measured force on DNA (8.4 kbp) tethered microspheres (silica, diameter 1.5 μm) distributed over a distance of more than a millimeter. The grey curve represents the force distribution expected from the size variation from microsphere to microsphere (σbead ≍ 6%). This variation is probably the limiting factor of the force homogeneity in our setup
Supplementary Figure 4 The stability of the AFS setup.
The measured end-to-end distance of a DNA molecule (10.1 kbp) to illustrate the stability of the AFS apparatus. The DNA molecule was tethered to a 4.5 μm polystyrene microsphere that was subjected to an acoustic force of 9 pN. No significant drifts were observed over a timescale of 16 hours.
Supplementary Figure 5 Selection criteria for single tethers.
XY scatter plots for a microsphere that was stuck (left panel), double tethered (middle panel) and single tethered (right panel). Stuck microspheres were discarded by their low RMS value and microspheres tethered to multiple molecules were discarded by their asymmetric motion (high symmetry value).
Supplementary Figure 6 Typical bond-rupture measurement.
A typical bond-rupture measurement of a DIG/anti-DIG bond. This measurement was performed for thousands of microspheres in parallel. At the beginning of the measurement no force was applied to determine the RMS and symmetry value of each tether in order to discriminate between single tethered microspheres and microspheres that were stuck or tethered to multiple molecules (Supplementary Fig. 5). To calibrate forces acting on individual microspheres, a constant voltage was applied for several minutes and the power spectrum of the quenched x, y motion was analyzed for each microsphere independently. Finally, a linear force ramp (~0.02 pN/s ) was applied. After the Dig/anti-Dig bond breaks, the microsphere leaves the selected tracking region of interest, leading to an increased value of Δ Template (equation (8), Online Methods ) yielding a random tracked positions. Note that, in these experiments, the positions were not corrected for drift.
Supplementary Figure 7 Sample heating.
Temperature measurement of the sample using a Platinum thermocouple that is embedded in the upper glass layer close to the piezo. The left panel shows that the heating is approximately 3℃ if 4 Vpp is applied for several minutes. Using a heatsink, a block of aluminum that is connected to the flow cell next to the piezo, decreases heating of the sample by a third. A more efficient heatsink will probably reduce the heating further. The right panel shows that we measure almost no heating for fast force ramps to high voltages (20 Vpp). Note that heating can be decreased if silica microspheres are used instead of polystyrene spheres since less than half the power is needed to achieve the same forces. Another option to decrease the amount of input power is simply using larger microspheres.
Supplementary Figure 8 Influence of layer thicknesses.
Simulated frequencies of three acoustic resonances of the probe chamber and the corresponding forces at the upper glass/water interface per electrical input power of the transducer when varying the thickness of an individual layer. A positive force is directed towards the fluid layer and is calculated for 4.5 μm polystyrene microspheres. The dashed lines indicate the actual thicknesses for our device, which are also the values used in the simulation for the layers that are not varied. The results show that varying the fluid layer thickness has the largest influence on the force.
Supplementary Figure 9 Effect of blurring and aliasing.
Theoretical power spectra calculated for a typical, tethered microsphere (diameter 4.5 µm; DNA length 8.4 kbp) that is subjected to a force of 5 pN. For this frequency range motion blurring due to the finite integration time (15 ms) effectively suppresses the extra power due to aliasing. Therefore there is no significant difference between the power spectrum without blurring or aliasing effects (black line) and the power spectrum that takes both effects into account (blue line).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 1002 kb)
Supplementary Software
AFS software package containing both the tracking and analysis software (see “AFS software package” folder for the readme file). Additional hardware drivers used in our setup are included. (ZIP 11435 kb)
Rights and permissions
About this article
Cite this article
Sitters, G., Kamsma, D., Thalhammer, G. et al. Acoustic force spectroscopy. Nat Methods 12, 47–50 (2015). https://doi.org/10.1038/nmeth.3183
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3183
This article is cited by
-
Acoustic tweezers for high-throughput single-cell analysis
Nature Protocols (2023)
-
Acoustofluidics for biomedical applications
Nature Reviews Methods Primers (2022)
-
Low cost and massively parallel force spectroscopy with fluid loading on a chip
Nature Communications (2022)
-
Spatiotemporally controlled generation of NTPs for single-molecule studies
Nature Chemical Biology (2022)
-
Harmonic acoustics for dynamic and selective particle manipulation
Nature Materials (2022)