Abstract
The comprehensive reconstruction of cell lineages in complex multicellular organisms is a central goal of developmental biology. We present an open-source computational framework for the segmentation and tracking of cell nuclei with high accuracy and speed. We demonstrate its (i) generality by reconstructing cell lineages in four-dimensional, terabyte-sized image data sets of fruit fly, zebrafish and mouse embryos acquired with three types of fluorescence microscopes, (ii) scalability by analyzing advanced stages of development with up to 20,000 cells per time point at 26,000 cells min−1 on a single computer workstation and (iii) ease of use by adjusting only two parameters across all data sets and providing visualization and editing tools for efficient data curation. Our approach achieves on average 97.0% linkage accuracy across all species and imaging modalities. Using our system, we performed the first cell lineage reconstruction of early Drosophila melanogaster nervous system development, revealing neuroblast dynamics throughout an entire embryo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
20 August 2014
In the version of this article initially published online, equation (2) in the Online Methods, which describes the expression used to calculate the distance between the location of a nucleus (
 ) and the plane defined by the vertices of a triangle on the convex hull (
) and the plane defined by the vertices of a triangle on the convex hull (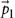 ,
,  and
and  ) was incorrect. The operations between the positions of the nucleus and triangle vertices (
) was incorrect. The operations between the positions of the nucleus and triangle vertices ( ,
, 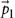 ,
,  and
and  ) were incorrectly shown as a scalar product. The correct operation is a subtraction. The operations between the pairwise differences of the positions of the triangle vertices (
) were incorrectly shown as a scalar product. The correct operation is a subtraction. The operations between the pairwise differences of the positions of the triangle vertices (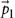 ,
,  and
and  ) were also incorrectly shown as a scalar product. The correct operation is a vector product. These errors have been corrected for the print, PDF and HTML versions of this article.
) were also incorrectly shown as a scalar product. The correct operation is a vector product. These errors have been corrected for the print, PDF and HTML versions of this article.
References
Megason, S.G. & Fraser, S.E. Imaging in systems biology. Cell 130, 784–795 (2007).
Khairy, K. & Keller, P.J. Reconstructing embryonic development. Genesis 49, 488–513 (2011).
Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H.K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008).
McMahon, A., Supatto, W., Fraser, S.E. & Stathopoulos, A. Dynamic analyses of Drosophila gastrulation provide insights into collective cell migration. Science 322, 1546–1550 (2008).
Fernandez, R. et al. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat. Methods 7, 547–553 (2010).
Bosveld, F. et al. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science 336, 724–727 (2012).
Lemon, W.C. & Keller, P.J. Live imaging of nervous system development and function using light-sheet microscopy. Mol. Reprod. Dev. 10.1002/mrd.22258 (2014).
Murray, J.I. et al. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Methods 5, 703–709 (2008).
Liu, X. et al. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell 139, 623–633 (2009).
Held, M. et al. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods 7, 747–754 (2010).
Amat, F. & Keller, P.J. Towards comprehensive cell lineage reconstructions in complex organisms using light-sheet microscopy. Dev. Growth Differ. 55, 563–578 (2013).
Trichas, G. et al. Multi-cellular rosettes in the mouse visceral endoderm facilitate the ordered migration of anterior visceral endoderm cells. PLoS Biol. 10, e1001256 (2012).
Xiong, F. et al. Specified neural progenitors sort to form sharp domains after noisy Shh signaling. Cell 153, 550–561 (2013).
Keller, P.J. Imaging morphogenesis: technological advances and biological insights. Science 340, 1234168 (2013).
Bao, Z. et al. Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103, 2707–2712 (2006).
Tomer, R., Khairy, K., Amat, F. & Keller, P.J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Swoger, J., Muzzopappa, M., Lopez-Schier, H. & Sharpe, J. 4D retrospective lineage tracing using SPIM for zebrafish organogenesis studies. J. Biophotonics 4, 122–134 (2011).
Giurumescu, C.A. et al. Quantitative semi-automated analysis of morphogenesis with single-cell resolution in complex embryos. Development 139, 4271–4279 (2012).
Olivier, N. et al. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science 329, 967–971 (2010).
Kausler, B.X. et al. A discrete chain graph model for 3D+t cell tracking with high misdetection robustness. ECCV 144–157 (2012).
Li, K. et al. Cell population tracking and lineage construction with spatiotemporal context. Med. Image Anal. 12, 546–566 (2008).
Smal, I. et al. Multiple object tracking in molecular bioimaging by Rao-Blackwellized marginal particle filtering. Med. Image Anal. 12, 764–777 (2008).
Jaqaman, K. et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008).
Amat, F., Myers, E.W. & Keller, P.J. Fast and robust optical flow for time-lapse microscopy using super-voxels. Bioinformatics 29, 373–380 (2013).
Skraba, P., Ovsjanikov, M., Chazal, F. & Guibas, L. Persistence-based segmentation of deformable shapes. Proc. CVPR 45–52 (2010).
Vincent, L. & Soille, P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 13, 583–598 (1991).
Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979).
Bishop, C.M. Pattern Recognition and Machine Learning (Springer, 2007).
Dasgupta, S. & Schulman, L.J. A two-round variant of EM for Gaussian mixtures. Proc. UCAI 152–159 (2000).
Milan, A., Schindler, K. & Roth, S. Challenges of ground truth evaluation of multi-target tracking. Proc. CVPR 735–742 (2013).
Saalfeld, S., Cardona, A., Hartenstein, V. & Tomančák, P. CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25, 1984–1986 (2009).
Cardona, A. Collaborative annotation toolkit for massive amounts of image data. CATMAID GitHub Repository https://github.com/acardona/CATMAID (2014).
Hartenstein, V. & Camposortega, J.A. Early neurogenesis in wild-type Drosophila melanogaster. Rouxs Arch. Dev. Biol. 193, 308–325 (1984).
Doe, C.Q. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development 116, 855–863 (1992).
Hartenstein, V., Younossi-Hartenstein, A. & Lekven, A. Delamination and division in the Drosophila neurectoderm: spatiotemporal pattern, cytoskeletal dynamics, and common control by neurogenic and segment polarity genes. Dev. Biol. 165, 480–499 (1994).
Broadus, J., et al. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech. Dev. 53, 393–402 (1995).
Wang, F., Dumstrei, K., Haag, T. & Hartenstein, V. The role of DE-cadherin during cellularization, germ layer formation and early neurogenesis in the Drosophila embryo. Dev. Biol. 270, 350–363 (2004).
Technau, G.M., Berger, C. & Urbach, R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev. Dyn. 235, 861–869 (2006).
Siegrist, S.E. & Doe, C.Q. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development 133, 529–536 (2006).
Bowman, S.K., Neumuller, R.A., Novatchkova, M., Du, Q. & Knoblich, J.A. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742 (2006).
Trichas, G., Begbie, J. & Srinivas, S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 6, 40 (2008).
Kuhn, H.W. The Hungarian method for the assignment problem. Nav. Res. Log. Quart. 2, 83–97 (1955).
Acknowledgements
We thank A. Cardona and the participants of the Janelia CATMAID hackathon for help with modifying the open source code of CATMAID; R. Chhetri and A. Pavlopoulos for invaluable contributions to ground truth annotations of the microscopy data sets; the Ilastik development team for help using Ilastik; M. Schroeder, H. Lacin, J. Truman and T. Lee for helpful discussion about the Drosophila nervous system; S. Srinivas and T. Watanabe (University of Oxford) for their generous help in exploring imaging assays for mouse embryonic development, helpful discussions about mouse embryo culturing and providing the CAG-TAG1 transgenic mouse strain; A. Denisin for her outstanding help developing SiMView live imaging assays; C. Akitake (Carl Zeiss) for her generous help executing the Lightsheet Z.1 experiments; S. Olenych and O. Selchow (Carl Zeiss) for supporting the Lightsheet Z.1 experiments; and C.-P. Heisenberg (Institute of Science and Technology Austria) for kindly providing the Tg(β-actin:H2B-mCherry) and Tg(β-actin:H2B-eGFP) zebrafish lines. This work was supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
P.J.K. and F.A. conceived of the research with input from E.W.M. F.A. developed the cell lineaging framework and performed the cell lineage reconstructions with input from P.J.K. and K.B. W.L. performed the Drosophila imaging experiments. D.P.M. curated and analyzed the reconstruction of early Drosophila nervous system development. K.M. performed the mouse imaging experiments. Y.W. and W.L. performed the zebrafish imaging experiments. F.A. and P.J.K. analyzed the data with input from K.B. F.A. and P.J.K. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Principles of persistence clustering with watershed
(a) One-dimensional example of a watershed. The function profile is segmented into three basins (red, blue, orange) by grouping convex segments associated with the same local minima.
(b) Graphical representation using a dendrogram of how persistence-based clustering (PBC) can establish a hierarchical order for merging the different regions generated by the watershed in (a). In order to merge two regions, the parameter τ needs to be set to a value higher than the difference between the function value at the contact point between the two regions and the higher of the two local minima. Every time two regions in the dendrogram are merged, the τ value for the next merge can change. Thus, the dendrogram needs to be calculated sequentially. For example, the merging of region 1 (red) and region {2,3} (blue plus orange) is associated with a τ equal to f1,{2,3} instead of f1,2, since region 3 has a lower local minimum than region 1.
Supplementary Figure 2 Graphical model for sequential Gaussian Mixture Models
Graphical model indicating conditional independence between all random variables present in our sequential Gaussian Mixture Model (GMM). Xt represents the observed intensity values in the image stack at time point t. Zt represents the hidden variables that assign each Xt to a mixture in the GMM. The grey box indicates that these two random variables are considered independent and identically distributed (i.i.d.). µt and Ʌt represent the mean matrix and precision matrix, respectively, for each mixture. πt represents the responsibility for each mixture. The sub-graph defined by variables Xt, Zt, µt, Ʌt and πt is the standard graphical model defining a GMM. We incorporate binary variable St for each mixture to define the probability that a nucleus is dividing. Finally, we can unroll the model to perform inference sequentially in time as a dynamic Bayesian network. Thus, values at time point t – 1 act as priors for time point t.
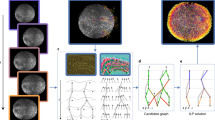
Supplementary Figure 3 Modeling of nuclei intensity profiles as Gaussian distributions
(a) Slices of an image stack in orthogonal planes xy (sliced orthogonally to detection axis) and xz (sliced parallel to detection axis) passing through the center of a segmented nucleus in a Drosophila dataset recorded with SiMView microscopy.
(b) Gaussian fits (black lines) to the projected intensity levels (green dots) of the stack in (a) along each of three main axes (marginal projections). The high accuracy of the fits in all three directions validates the assumption that nucleus intensity profiles can generally be well approximated by a Gaussian Mixture Model.
(c,e,g,i) Same as in (a), but for image slices obtained from a zebrafish data set recorded with SiMView microscopy (c), a mouse data set recorded with SiMView microscopy (e), a Drosophila data set recorded with a Carl Zeiss Lightsheet Z.1 light-sheet microscope (g) and a zebrafish data set recorded with a Carl Zeiss LSM 710 confocal microscope (i).
(d,f,h,j) Same as in (b), but for the image data shown in (c) (for (d)), (e) (for (f)), (g) (for (h)) and (i) (for (j)).
Supplementary Figure 4 Precision-recall curve for background detection module
Precision-recall curve (blue line) for the background classifier trained on spatio-temporal features (Supplementary Note 2). The size of the training set was 43,500 samples and the size of the test set was 29,000 samples. All samples were annotated using the CATMAID interface discussed in section “Visualization and manual curation of lineaging results” in the main text. The machine learning classifier is based on the RUSBoost implementation in Matlab with 600 classification trees as weak classifiers. Each tree is grown and then pruned such that each leaf contains a minimum of 20 training samples. In order to avoid losing real nuclei, the threshold for detecting trajectories with background objects (red line) should be set to a value higher than 0.6.
Supplementary Figure 5 Parameter sensitivity analysis
Parameter sensitivity analysis for the two tunable parameters of the processing pipeline, the image background intensity threshold and the PBC threshold τ for watershed agglomeration, with respect to Euclidean distance error metric (a,d), nearest neighbor (NN) normalized Euclidean distance error metric (b,e) and linkage error metric (c,f). Please see Supplementary Note 3 for details on these metrics.
Panels (a-c) show results for the Drosophila SiMViev dataset. In this scenario, the image background intensity threshold has a larger impact on accuracy than τ, owing to the high signal-to-noise ratio (SNR) of the data set. However, sensitivity with respect to this threshold is low enough to ensure close-to-optimal results for a wide range of parameters values.
Panels (d-f) show results for the zebrafish confocal dataset. In this scenario, τ has a larger impact on accuracy due to the lower SNR. While the ranges of close-to optimal values for image background intensity are quite different in (a-c) and (d-f), the range of close-to-optimal values for τ is almost identical. This observation is true for all datasets investigated in this study. Thus, for new datasets, we generally recommend using τ values between 5 and 15.
Supplementary Figure 6 Examples of segmentation and tracking errors
Orthogonal optical slices at different locations in the SiMView recording, each centered on a nucleus representing a different type of error in the automatic segmentation and tracking pipeline. The automated segmentation result is indicated using green outlines.
(a,b) Linkage errors due to large displacements of cell nuclei between consecutive time points. Magenta outlines indicate the respective correct solutions.
(c) Under-segmentation along the optical detection axis, owing to low image contrast.
(d) Under-segmentation within the image plane, owing to proximity of cell nuclei.
(e) Over-segmentation owing to uneven staining of the nuclear fluorescent label. The algorithm interprets the image content as a cell division.
(f) Over-segmentation owing to uneven staining of the nuclear fluorescent label, and subsequent error recovery. The algorithm recovers from an error similar to the one shown in panel (e) by ending one of the two tracks and propagating the other one in time with the correct segmentation solution. Thus, errors do not accumulate over time in the sequential propagation of the Gaussian mixture model.
Scale bar, 10 µm.
Supplementary Figure 7 Error analysis as a function of developmental stage, signal-to-noise ratio, cell density and imaging depth
Histograms displaying how different factors affect lineage reconstruction accuracy of the automated segmentation and tracking method presented in this study. All histograms were extracted from n = 5,331 linkage annotations in the SiMView time-lapse recording of Drosophila embryogenesis. The ranges of the plots span nearly the full physical limits of parameters measured in this data set, which are as follows: the nuclei nearest neighbor distance ranges from 4 µm to 21 µm, the nuclei distance to the center of the embryo (with a diameter of ~200 µm) ranges from 0 to 106 µm, centroid displacements range from 0 µm to 44 µm between consecutive time points, and the 90th percentile of the local image contrast is 33.
(a) The larger the distance between adjacent nuclei, the higher the quality of the cell lineage reconstruction.
(b) The shorter the path traveled by the fluorescent signal to the detection objective, the higher the image quality and the better the cell lineage reconstruction.
(c) The quality of the cell lineage reconstruction is independent of the nucleus displacement between two time points, as long as this displacement is not too large. This is the main assumption in our framework and for very large displacements the method breaks down (note that the scale of the vertical axis is substantially different compared to the other plots). Optical flow techniques can extend this range substantially and enable successful application of the automated segmentation and tracking framework also in the presence of larger displacements, if necessary. However, such an extension was not required for any of the data examples presented in this study.
(d) The higher the local image contrast (ratio of nucleus brightness versus background level), the better the cell lineage reconstruction.
Supplementary Figure 8 Disagreement between annotators
Comparison of Euclidean distance accuracy between different annotators for time point 400 of the SiMView time-lapse recording of zebrafish embryonic development (Supplementary Video 19).
(a) Histogram of Euclidean distances between centroid locations annotated by human annotator 1 (reference) and those obtained automatically with our automated cell lineage reconstruction framework (Supplementary Table 2). The average centroid distance (i.e. accuracy of the automated framework) is 1.29 µm (n = 734).
(b) Histogram of Euclidean distances between centroid locations annotated by human annotator 1 (reference) versus human annotator 2. Both annotators independently marked the centroid locations for the same set of cells, using the CATMAID interface and without knowledge of the other user’s annotation. The average centroid distance (i.e. inter-user accuracy) is 0.89 µm (n = 200).
Supplementary Figure 9 Spatio-temporal clustering of tracking and segmentation errors
Analysis of co-localization in space and time of all errors (4,982) found in the automated reconstruction of cell lineages described in Fig. 4. To obtain this histogram we constructed a graph where each error defines one node. A node is connected by an edge to another node from the same time point if (and only if) this second node is one of the first node’s four nearest neighbors. Two nodes belonging to different time points are connected by an edge if (and only if) they belong to the same lineage in the automatic reconstruction and are less than two time points apart. The figure shows the histogram of sizes of the different connected components in the resulting graph.
Supplementary Figure 10 Image volume read time overhead in cell lineage reconstructions
Time required to read a single three-dimensional image stack for each of the data sets discussed in the main text. For each time point of a time-lapse data set, the respective image stack is read twice: once for the initial hierarchical segmentation and once in the sequential GMM tracking module. All image data were stored using lossless compression in three-dimensional JPEG2000 format using libraries from the PICTools Medical software package. Read time increases quadratically with image size and was measured on a processing workstation equipped with two Intel Xeon E5-2687W CPUs, six Seagate Savvio 10K.5 ST9900805SS hard disks combined in a RAID-6 array and an Intel RMS25CB080 RAID module (Online Methods).
Supplementary Figure 11 Cell lineage reconstructions in the early zebrafish embryo
Proof-of-principle automated reconstruction and manual curation of cell lineages in the early zebrafish embryo. The underlying automated reconstructions are based on the data set shown in Supplementary Video 19 and are visualized in Supplementary Videos 20 and 21. The entire data set comprises 10.7 million data points (with one data point corresponding to the positions and dimensions of a cell nucleus at one time point). Manual data curation of the automatically reconstructed cell tracks was performed at a rate of 1,019 data points per hour.
(a) Spinal cord cells were manually identified in the automated cell lineage reconstruction, based on their spatial location at time point 720 in the zebrafish recording shown in Supplementary Videos 19-21. Using the data curation and annotation interface provided as Supplementary Software 2, the tracks of these cells were then curated backwards in time for the time interval 85-720 (10.6 hours of live imaging data, recorded at 21.5°C). When encountering a cell division, the respective other daughter cell was followed forwards in time and its track was fully curated as well. The panels in (a) visualize the resulting cell lineage reconstruction at three different time points, with yellow spheres indicating cell positions at the given time point and colored lines indicating the tracking information up to the respective time point.
(b) As in (a), but for a set of cells located in the anterior neural plate.
(c,d) Enlarged view of the boxed regions shown in (a) and (b) at time point 140, with annotation labels for the four spinal cord precursors (c) and four anterior neural plate precursors (d).
(e) Cell lineage tree representation of the cell lineage reconstructions visualized in (a) an (b). In addition to the start and end points of the reconstructed time interval, these trees include annotations of cell division time points. Each precursor divided at least once within the 10.6-hour imaging interval.
SCP = Spinal cord precursor, NPP = Anterior neural plate precursor.
Scale bars, 100 µm (a,b), 20 µm (c).
Supplementary Figure 12 Spatial maps of annotated neuroblasts
(a) Spatial map of the neuroblast array from our reconstruction of early Drosophila embryonic nervous system development at 4.4 h AEL, shortly after neuroblast internalization, using a color code for the manual neuroblast type annotation. Neuroblast coordinates in this map correspond to those shown in Fig. 5e and Supplementary Fig. 15. The manual annotation of neuroblast types was performed on the basis of relative positional information within the stereotypic neuroblast array, as previously described.
(b) Spatial map of the neuroblast array at 5.0 h AEL, approximately half an hour after neuroblast internalization. The color code was propagated from neuroblasts in panel (a) to the corresponding neuroblasts at this later time point, using the tracking information from the curated cell lineage reconstruction shown in Supplementary Fig. 13. Neighbor relationships are largely preserved over this time interval.
Supplementary Figure 13 Cell lineage reconstruction of the early Drosophila embryonic nervous system
Cell lineage tree for all neuroblasts tracked in our reconstruction of early Drosophila embryonic nervous system development (Fig. 5, Supplementary Videos 24-28). Green circles indicate lineage origins in the blastoderm, red circles indicate cell division events and blue circles indicate the end time point of cell tracks for the respective neuroblasts or ganglion mother cells. All neuroblasts identified by manual inspection are annotated at the end of the respective lineage branch.
Supplementary Figure 14 Neuroblast division angles
Local coordinate system used to analyze neuroblast division angles (ψ, η). The angles ψ, η define a spherical coordinate system. The origin corresponds to the center of the ganglion mother cell. The north pole is defined by the normal vector to the surface of the embryo at the point closest to the ganglion mother cell. The division angle η defines the deviation of the surface normal from the vector connecting ganglion mother cell and neuroblast. The division angle ψ defines the orientation of the division axis relative to the anerior-posterior and medio-lateral axes of the embryo. ψ = 0° corresponds to the direction along the germ band facing the anterior end of the embryo, and ψ = 270° corresponds to a medial direction.
Supplementary Figure 15 Morphodynamic measurements using neuroblast trajectories
(a) Overview of features of dynamic cell behavior measured for all neural precursors analyzed in the reconstruction of early Drosophila embryonic nervous system development (Fig. 5, Supplementary Videos 24-28). This overview figure includes the four features shown in the main text (Fig. 5e). Each feature is represented in the neuroblast array at 4.4 h AEL using an individual color code. These features were used for the prediction of neuroblast cell types, shown in Fig. 6e,f.
(b) Overview of bilateral symmetry analysis for the features of dynamic cell behavior measured for all neural precursors analyzed in the reconstruction of early Drosophila embryonic nervous system development. This overview figure includes the plot shown in the main text (Fig. 6b). Neuroblasts to the left of the midline are represented by red lines, neuroblasts to the right of the midline by blue lines. Continuity of the plots was achieved by using a Gaussian-weighted average along the midline to convert the discrete neuroblast data points to a continuous graph. The sigma of the Gaussian was set to 0.1 rad. The level of feature correlation in the left and right halves of the neuroblast array is indicated in the form of an R2 score above each plot.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Table 1–6, and Supplementary Notes 1–5 (PDF 3096 kb)
Supplementary Software 1
Automated modules of the cell lineaging framework (ZIP 227784 kb)
Supplementary Software 2
Modified CATMAID module for visualizing image and cell lineage data, manually curating cell lineage data and annotating cell lineage reconstructions (ZIP 19870 kb)
Supplementary Data 1
Cell lineage reconstruction of early Drosophila embryonic nervous system development (ZIP 1139 kb)
SiMView imaging of Drosophila embryogenesis
Simultaneous multi-view imaging of Drosophila embryonic development, using an embryo homozygous for the nuclear label Histone 2A-mRFP (w-; P{w[+mC]=His2Av-mRFP1}; +, stock number 23560 from the Bloomington Drosophila Stock Center). The embryo was recorded in 30-second intervals over a period of 24 hours, starting at 3 h AEL. The first 551 time points, which cover the full time period analyzed in the automated cell lineage reconstruction, are included in this video. The complete data set consists of 515 gigabytes of image data. The video shows separate maximum-intensity projections of the dorsal and ventral halves of the embryo, based on the fused and background-corrected three-dimensional image stacks. The image frame size was down-sampled to reduce video size. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 32052 kb)
Automated segmentation and tracking in SiMView data set of Drosophila embryogenesis (gradient color code)
Automated computational cell lineage reconstruction of the image data set shown in Supplementary Video 1. Each circle represents one cell nucleus. The tails of the circles (solid lines) indicate the history of object positions for the past ten time points. The color scheme was initialized in the first frame using a color gradient from anterior to posterior, using different colors on the dorsal and ventral sides and ensuring continuity in color space at the anterior and posterior ends of the embryo (see color bar in Figure 2a). After this initial color assignment, the color information was propagated in time using the tracking information, thus providing a color-coded single-cell resolution fate map. The accuracy of this automated procedure is quantified in Figure 2e. Some cell nucleus detections correspond to background objects, arising from autofluorescence and limitations in image quality. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 46019 kb)
Automated segmentation and tracking in SiMView data set of Drosophila embryogenesis (random color code)
Visualization as in Supplementary Video 2, but using a random color code to initialize the first video frame. As in Supplementary Video 2, the color information was then propagated in time using the tracking information. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 45938 kb)
Automated segmentation and tracking in SiMView data set of Drosophila embryogenesis (rotating embryo, color gradient)
Visualization as in Supplementary Video 2, but using a rotating view of the embryo instead of split static views of the dorsal and ventral halves. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 38785 kb)
Image volume slicing sequence for time point 30 of the reconstructed SiMView data set of Drosophila embryogenesis
Three-dimensional image stack from the SiMView recording of the nuclei-labeled Drosophila embryo shown in Supplementary Video 1, superimposed with the corresponding supervoxels of the low-level over-segmentation (magenta) and the ellipsoids for all automatically reconstructed objects (green). The video shows the three-dimensional image data plane per plane for time point 30 of the image data set, shortly after the onset of gastrulation (3.2 h AEL). The image frame size was down-sampled to reduce video size. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 22889 kb)
Image volume slicing sequence for time point 180 of the reconstructed SiMView data set of Drosophila embryogenesis
As in Supplementary Video 5, but for a time point later in development (time point 180, corresponding to 4.4 h AEL). The identification and annotation of neuroblasts for the reconstruction of early Drosophila embryonic nervous system development (Supplementary Videos 24-28) was performed at this time point. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 26868 kb)
Lightsheet Z.1 imaging of Drosophila embryogenesis
Light-sheet-based imaging of Drosophila embryonic development with a Carl Zeiss Lightsheet Z.1 microscope, using an embryo homozygous for the nuclear label Histone 2A-mRFP (w-; P{w[+mC]=His2Av-mRFP1}; +, stock number 23560 from the Bloomington Drosophila Stock Center). The embryo was recorded in 30-second intervals over a period of 11 hours, starting at 2 h AEL. The first 387 time points, which cover the full time period analyzed in the automated cell lineage reconstruction, are included in this video. The complete data set consists of 595 gigabytes of image data. The video shows maximum-intensity projections of the ventral half of the embryo, based on the background-corrected three-dimensional image stacks. Note that, in contrast to the SiMView recording, which allows quantitatively accurate fusion of all views to produce a near-complete data set of the developing embryo, the Lightsheet Z.1 microscope only allows sequential multi-view imaging (and, thus, physical rotation of the embryo is needed) to capture the dorsal and ventral halves of the embryo. To avoid image fusion artifacts, the data sets representing the dorsal and ventral halves of the embryo were therefore not fused and only cell lineages in the ventral half of the embryo were reconstructed. Image frame size was down-sampled to reduce video size. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 15117 kb)
Automated segmentation and tracking in Lightsheet Z.1 data set of Drosophila embryogenesis (gradient color code)
Automated computational cell lineage reconstruction of the image data set shown in Supplementary Video 7. Each circle represents one cell nucleus. The tails of the circles (solid lines) indicate the history of object positions for the past ten time points. The color scheme was initialized in the first frame using a color gradient from anterior to posterior. After this initial color assignment, the color information was propagated in time using the tracking information, thus providing a color-coded single-cell resolution fate map. Some cell nucleus detections correspond to background objects, arising from autofluorescence and limitations in image quality. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 30407 kb)
Automated segmentation and tracking in Lightsheet Z.1 data set of Drosophila embryogenesis (random color code)
Visualization as in Supplementary Video 8, but using a random color code to initialize the first video frame. As in Supplementary Video 8, the color information was then propagated in time using the tracking information. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 30545 kb)
Image volume slicing sequence for time point 90 of the reconstructed Lightsheet Z.1 data set of Drosophila embryogenesis
Three-dimensional image stack from the Lightsheet Z.1 recording of the nuclei-labeled Drosophila embryo shown in Supplementary Video 7, superimposed with the corresponding supervoxels of the low-level over-segmentation (magenta) and the ellipsoids for all automatically reconstructed objects (green). The video shows the three-dimensional image data plane per plane for time point 90 of the image data set, at the onset of gastrulation (3 h AEL). The image frame size was down-sampled to reduce video size. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 10594 kb)
Image volume slicing sequence for time point 240 of the reconstructed Lightsheet Z.1 data set of Drosophila embryogenesis
As in Supplementary Video 10, but for a time point later in development (time point 240, corresponding to 4.3 h AEL). Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 11888 kb)
Confocal imaging of zebrafish embryogenesis
Confocal microscopy recording of zebrafish embryonic development with a Carl Zeiss LSM 710 laser-scanning confocal microscope, using an embryo heterozygous for the fluorescent nuclear label H2B-mCherry. The embryo was recorded in 120-second intervals over a period of 3.4 hours. The data set consists of 9.4 gigabytes of image data. The video shows maximum-intensity projections with a view of the animal pole of the embryo. The image frame size was down-sampled to reduce video size. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 11800 kb)
Automated segmentation and tracking in confocal data set of zebrafish embryogenesis (gradient color code)
Automated computational cell lineage reconstruction of the image data set shown in Supplementary Video 12. Each circle represents one cell nucleus. The tails of the circles (solid lines) indicate the history of object positions for the past ten time points. The color scheme was initialized in the first frame using a radially-symmetrical color gradient from the animal pole to the periphery of the blastoderm. After this initial color assignment, the color information was propagated in time using the tracking information, thus providing a color-coded single-cell resolution fate map. Some cell nucleus detections correspond to background objects, arising from autofluorescence and limitations in image quality. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 16421 kb)
Automated segmentation and tracking in confocal data set of zebrafish embryogenesis (random color code)
Visualization as in Supplementary Video 13, but using a random color code to initialize the first video frame. As in Supplementary Video 13, the color information was then propagated in time using the tracking information. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 16215 kb)
Image volume slicing sequence for time point 60 of the reconstructed confocal data set of zebrafish embryogenesis
Three-dimensional image stack from the LSM 710 recording of the nuclei-labeled zebrafish embryo shown in Supplementary Video 12, superimposed with the corresponding supervoxels of the low-level over-segmentation (magenta) and the ellipsoids for all automatically reconstructed objects (green). The video shows the three-dimensional image data plane per plane for time point 60 of the image data set. The video frame size is the original size of the image data. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 13398 kb)
SiMView imaging of mouse embryogenesis
Simultaneous multi-view imaging of mouse embryonic development, using an embryo expressing Histone2B-eGFP in all nuclei. The E6.25 embryo was recorded in 5-minute intervals over a period of 2 hours. The data set consists of 8.4 gigabytes of image data. The video shows maximum-intensity projections of the embryo, based on the fused and background-corrected three-dimensional image stacks. The video frame size is the original size of the image data. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 5776 kb)
Automated segmentation and tracking in SiMView data set of mouse embryogenesis (random color code)
Automated computational cell lineage reconstruction of the image data set shown in Supplementary Video 16. Each circle represents one cell nucleus. The tails of the circles (solid lines) indicate the history of object positions for the past ten time points. The color scheme was initialized in the first frame using a random color code. After this initial color assignment, the color information was propagated in time using the tracking information. Some cell nucleus detections correspond to background objects, arising from autofluorescence and limitations in image quality. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 9418 kb)
Image volume slicing sequence for time point 24 of the reconstructed SiMView data set of mouse embryogenesis
Three-dimensional image stack from the SiMView recording of the nuclei-labeled mouse embryo shown in Supplementary Video 16, superimposed with the corresponding supervoxels of the low-level over-segmentation (magenta) and the ellipsoids for all automatically reconstructed objects (green). The video shows the three-dimensional image data plane per plane for time point 24 of the image data set. The video frame size is the original size of the image data. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 13931 kb)
SiMView imaging of zebrafish embryogenesis
Simultaneous multi-view imaging of zebrafish embryonic development, using an embryo heterozygous for the nuclear label H2B-GFP, expressed under the control of the beta-actin promoter. The embryo was recorded in 60-second intervals over a period of 18 hours, starting at the sphere stage at 6 hours post fertilization. The first 541 time points are included in this video. The complete data set consists of 1.7 terabytes of image data. The video shows separate maximum-intensity projections of the animal and vegetal halves of the embryo, based on the fused and background-corrected three-dimensional image stacks. The image frame size was down-sampled to reduce video size. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 32598 kb)
Automated segmentation and tracking in SiMView data set of zebrafish embryogenesis (gradient color code)
Automated computational cell lineage reconstruction of the image data set shown in Supplementary Video 19. Each circle represents one cell nucleus. The tails of the circles (solid lines) indicate the history of object positions for the past ten time points. The color scheme was initialized in the first frame using a radially-symmetrical color gradient from the animal pole to the periphery of the blastoderm. After this initial color assignment, the color information was propagated in time using the tracking information, thus providing a color-coded single-cell resolution fate map. Some cell nucleus detections correspond to background objects, arising from autofluorescence and limitations in image quality. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 73705 kb)
Automated segmentation and tracking in SiMView data set of zebrafish embryogenesis (random color code)
Visualization as in Supplementary Video 20, but using a random color code to initialize the first video frame. As in Supplementary Video 20, the color information was then propagated in time using the tracking information. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 73618 kb)
Image volume slicing sequence for time point 100 of the reconstructed SiMView data set of zebrafish embryogenesis
Three-dimensional image stack from the SiMView recording of the nuclei-labeled zebrafish embryo shown in Supplementary Video 19, superimposed with the corresponding supervoxels of the low-level over-segmentation (magenta) and the ellipsoids for all automatically reconstructed objects (green). The video shows the three-dimensional image data plane per plane for time point 100 of the image data set (7.7. hours post fertilization, imaging at 21.5°C). The image frame size was down-sampled to reduce video size. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 35700 kb)
Image volume slicing sequence for time point 400 of the reconstructed SiMView data set of zebrafish embryogenesis
As in Supplementary Video 22, but for a time point later in development (time point 400, corresponding to 12.7 hours post fertilization; imaging at 21.5°C). Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 27686 kb)
Neuroblast identification in the SiMView data set of Drosophila embryogenesis
Rotating maximum-intensity projection of the three-dimensional SiMView image stack of the nuclei-labeled Drosophila embryo shown in Supplementary Video 1 (at time point 180, corresponding to 4.4 h AEL), superimposed with green spheres marking the location of neuroblasts identified in the image data. Neuroblast identification was performed based on morphological appearance and cell position in the embryo. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 25658 kb)
Movements and divisions of neural precursors in the early Drosophila embryonic nervous system (with SiMView data)
Ventral and lateral maximum-intensity projections of the SiMView time-lapse recording of the nuclei-labeled Drosophila embryo shown in Supplementary Video 1, superimposed with green spheres marking the location, movements and divisions of neural precursors from the blastoderm stage up to 5 h AEL The tails indicate the history of cell positions for the past ten time points, using a color code that gradually transitions from purple to white as a function of time. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 30593 kb)
Movements and divisions of neural precursors in the early Drosophila embryonic nervous system (static view)
Visualization as in Supplementary Video 2, but using a cell type specific color code obtained from the cell lineage reconstruction of early Drosophila embryonic nervous system development. Neural precursors are color-coded based on neuroblast type (see Supplementary Figure 12 for color correspondence) and all other cells are shown in grey. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 25094 kb)
Movements and divisions of neural precursors in the early Drosophila embryonic nervous system (rotating view)
Visualization as in Supplementary Video 26, but using a rotating view of the embryo instead of split static views of the dorsal and ventral halves. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 25346 kb)
Cell lineage reconstruction of early Drosophila embryonic nervous system development
Rotating view of neural precursor cell tracks obtained from the cell lineage reconstruction of early Drosophila embryonic nervous system development. The tracks are represented by solid lines, using a color code that indicates time (purple to white: from the blastoderm stage to the end point of the reconstruction at 5 h AEL). Cell locations at the end point of the reconstruction are marked by green spheres. Anterior is to the left, posterior to the right. Note: The DivX codec required for video playback is freely available at http://www.divx.com/downloads/divx/1 (AVI 35773 kb)
Rights and permissions
About this article
Cite this article
Amat, F., Lemon, W., Mossing, D. et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat Methods 11, 951–958 (2014). https://doi.org/10.1038/nmeth.3036
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3036
This article is cited by
-
Image restoration of degraded time-lapse microscopy data mediated by near-infrared imaging
Nature Methods (2024)
-
Tutorial: a beginner’s guide to building a representative model of dynamical systems using the adjoint method
Communications Physics (2024)
-
An archetype and scaling of developmental tissue dynamics across species
Nature Communications (2023)
-
HyU: Hybrid Unmixing for longitudinal in vivo imaging of low signal-to-noise fluorescence
Nature Methods (2023)
-
Automated reconstruction of whole-embryo cell lineages by learning from sparse annotations
Nature Biotechnology (2023)



 ) and the plane defined by the vertices of a triangle on the convex hull (
) and the plane defined by the vertices of a triangle on the convex hull (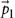 ,
,  and
and  ) was incorrect. The operations between the positions of the nucleus and triangle vertices (
) was incorrect. The operations between the positions of the nucleus and triangle vertices (