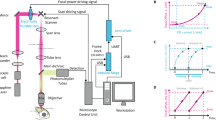
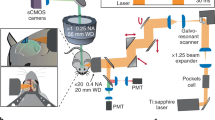
Abstract
High-speed, large-scale three-dimensional (3D) imaging of neuronal activity poses a major challenge in neuroscience. Here we demonstrate simultaneous functional imaging of neuronal activity at single-neuron resolution in an entire Caenorhabditis elegans and in larval zebrafish brain. Our technique captures the dynamics of spiking neurons in volumes of ∼700 μm × 700 μm × 200 μm at 20 Hz. Its simplicity makes it an attractive tool for high-speed volumetric calcium imaging.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Alivisatos, A.P. et al. Neuron 74, 970–974 (2012).
Marblestone, A.H. et al. Front. Comput. Neurosci. 7, 137 (2013).
Stosiek, C., Garaschuk, O., Holthoff, K. & Konnerth, A. Proc. Natl. Acad. Sci. USA 100, 7319–7324 (2003).
Chen, T.-W. et al. Nature 499, 295–300 (2013).
Schrödel, T., Prevedel, R., Aumayr, K., Zimmer, M. & Vaziri, A. Nat. Methods 10, 1013–1020 (2013).
Ahrens, M.B., Orger, M.B., Robson, D.N., Li, J.M. & Keller, P.J. Nat. Methods 10, 413–420 (2013).
Panier, T. et al. Front. Neural Circuits 7, 65 (2013).
Duemani Reddy, G., Kelleher, K., Fink, R. & Saggau, P. Nat. Neurosci. 11, 713–720 (2008).
Grewe, B.F., Langer, D., Kasper, H., Kampa, B.M. & Helmchen, F. Nat. Methods 7, 399–405 (2010).
Cheng, A., Gonçalves, J.T., Golshani, P., Arisaka, K. & Portera-Cailliau, C. Nat. Methods 8, 139–142 (2011).
Abrahamsson, S. et al. Nat. Methods 10, 60–63 (2013).
Levoy, M., Ng, R., Adams, A., Footer, M. & Horowitz, M. ACM Trans. Graph. 25, 924–934 (2006).
Levoy, M., Zhang, Z. & McDowall, I. J. Microsc. 235, 144–162 (2009).
Quirin, S., Peterka, D.S. & Yuste, R. Opt. Express 21, 16007–16021 (2013).
Agard, D.A. Annu. Rev. Biophys. Bioeng. 13, 191–219 (1984).
Broxton, M. et al. Opt. Express 21, 25418–25439 (2013).
Faumont, S. et al. PLoS ONE 6, e24666 (2011).
Kawano, T. et al. Neuron 72, 572–586 (2011).
Chalfie, M. et al. J. Neurosci. 5, 956–964 (1985).
Mukamel, E.A., Nimmerjahn, A. & Schnitzer, M.J. Neuron 63, 747–760 (2009).
Friedrich, R.W. & Korsching, S.I. Neuron 18, 737–752 (1997).
Renninger, S.L. & Orger, M.B. Methods 62, 255–267 (2013).
Jetti, S.K., Vendrell-Llopis, N. & Yaksi, E. Curr. Biol. 24, 434–439 (2014).
Akerboom, J. et al. Front. Mol. Neurosci. 6, 2 (2013).
Ntziachristos, V. Nat. Methods 7, 603–614 (2010).
Cold Spring Harbor Laboratory. Hermaphrodite Handbook. WormAtlas (ed. Herndon, L.A.) http://www.wormatlas.org/hermaphrodite/hermaphroditehomepage.htm (2014; accessed 20 March 2014).
Kak, A.C. & Slaney, M. Principles of Computerized Tomographic Imaging (Society of Industrial and Applied Mathematics, 2001).
Gu, M. Advanced Optical Imaging Theory (Springer, 1999).
Acknowledgements
We thank T. Müller, P. Pasierbek, P. Forai, H. Kaplan, M. Molodtsov, K. Tessmar-Raible, F. Schlumm and Olympus Inc. for technical support and loan of equipment, as well as H. Baier (Max Planck Institute of Neurobiology) and M. Orger (Champalimaud) for sharing zebrafish lines. We thank L. Page for providing early funding for the project and D. Dalrymple for helping catalyze connections. The computational results presented have been achieved in part using the Vienna Scientific Cluster (VSC). This work was supported by the VIPS Program of the Austrian Federal Ministry of Science and Research and the City of Vienna as well as the European Commission (Marie Curie, FP7-PEOPLE-2011-IIF) (R.P.); a Samsung Scholarship (Y.-G.Y.); a US National Science Foundation (NSF) Graduate Fellowship (N.P.); the Allen Institute for Brain Science, the MIT Media Lab, the MIT McGovern Institute, US National Institutes of Health (NIH) 1R01EY023173, the MIT Synthetic Intelligence Project, the Institution of Engineering and Technology (IET) Harvey Prize, NSF CBET 1053233, the New York Stem Cell Foundation–Robertson Award, NSF CBET 1344219, NIH 1DP1NS087724, Google, the NSF Center for Brains, Minds and Machines at MIT, and Jeremy and Joyce Wertheimer (E.S.B.); the Vienna Science and Technology Fund (WWTF) project VRG10-11, Human Frontiers Science Program Project RGP0041/2012, Research Platform Quantum Phenomena and Nanoscale Biological Systems (QuNaBioS) (A.V.); and the European Community's Seventh Framework Programme/ERC no. 281869 (M.Z. and T.S.). The Institute of Molecular Pathology is funded by Boehringer Ingelheim.
Author information
Authors and Affiliations
Contributions
R.P. designed microlenses, built the imaging system and performed experiments together with M.H. Y.-G.Y. designed and wrote 3D-deconvolution software with contributions from G.W. under the guidance of R.R. R.P. and M.H. refined and rebuilt the imaging system and analyzed data together with Y.-G.Y. N.P. implemented and tested the LFDM prototype. T.S. generated transgenic animals, provided microfluidic devices and performed cell identifications under the guidance of M.Z. S.K. wrote analysis software. E.S.B. and A.V. conceived of and led project. R.P., Y.-G.Y. and A.V. wrote the manuscript, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Notes 1 and 2 (PDF 6322 kb)
Supplementary Software
Software for 3D volume reconstruction from light-field images The software for 3D volume reconstruction from light-field images is available as Supplementary Software. It is written in MATLAB and requires a workstation with MATLAB2013a or later version and at least 32GB of RAM. A user manual for the software (LFM_Software_readme.pdf) is included in the .zip file. (ZIP 7047 kb)
Whole animal Ca2+-imaging of C. elegans
Maximum intensity projection of 14 z-planes at 2 μm distance of a Punc-31::NLS-GCaMP5K worm. Shown are 200 seconds of recording of basal activity at 5 Hz volume rate (1000 frames in total). Video frame rate is 100 frames per second, which equates to 20 seconds in real time in the video (i.e. playback speed 20x). See also Fig. 2a-c. (AVI 7320 kb)
Whole animal Ca2+-imaging of C. elegans
Maximum intensity projection of 14 z-planes at 2 μm distance of a Punc-31::NLS-GCaMP5K worm. Shown are 200 seconds of recording of basal activity at 5 Hz volume rate. Video frame rate is 100 frames per second, which equates to 20 seconds in real time in the video (i.e. playback speed 20x). See also Fig. 2d. (AVI 9412 kb)
High-speed Ca2+-imaging of unrestrained C. elegans at 50 Hz
Maximum intensity projection of 12 z-planes at 2 μm distance of a Punc-31::NLS-GCaMP5K worm. Shown are 15 seconds of recording of basal activity of freely-moving worms at 50 Hz volume rate. Exposure time for individual volumes was 20 ms. Video frame rate is 50 frames per second, which equates to 50 volumes per second in the video (i.e. playback speed 1x – real time). (AVI 9533 kb)
Brain-wide Ca2+-imaging of C. elegans
Maximum intensity projection of 15 z-planes at 2 μm distance of a Punc-31::NLS-GCaMP5K worm. Shown are 200 seconds of recording of basal activity in the head ganglia at 5 Hz volume rate. Video frame rate is 100 frames per second, which equates to 20 seconds in real time in the video (i.e. playback speed 20x). See also Fig. 2e-f. (AVI 9673 kb)
Brain-wide Ca2+-imaging of C. elegans during chemosensory stimulation
Maximum intensity projection of 7 z-planes at 2 μm distance of a of a Punc-31::NLS-GCaMP5K worm during chemosensory stimulation. Shown are 240 seconds of recording of basal activity at 5 Hz volume rate. Video frame rate is 50 frames per second, which equates to 24 seconds in real time in the video (i.e. playback speed 10x). Also see Supplementary Fig. 3. (AVI 11061 kb)
Brain-wide Ca2+-imaging of zebrafish larvae
Maximum intensity projection of 51 z-planes at 4 μm distance of a 5 dpf - HuC:GCaMP5G zebrafish larvae. Shown are 240 seconds of recording of activity at 20 Hz volume rate. Playback speed of video is 10x. At ∼15 sec (∼1.5 sec in the video), decomposed fish water is supplied manually to the fish chamber in order to evoke activity in the olfactory system. Also see Fig. 3 in the main manuscript. (MP4 3748 kb)
Source data
Rights and permissions
About this article
Cite this article
Prevedel, R., Yoon, YG., Hoffmann, M. et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat Methods 11, 727–730 (2014). https://doi.org/10.1038/nmeth.2964
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2964
This article is cited by
-
Inferring how animals deform improves cell tracking
Nature Methods (2024)
-
Automated neuron tracking inside moving and deforming C. elegans using deep learning and targeted augmentation
Nature Methods (2024)
-
The spatial and temporal structure of neural activity across the fly brain
Nature Communications (2023)
-
Video-rate 3D imaging of living cells using Fourier view-channel-depth light field microscopy
Communications Biology (2023)
-
Mesoscale volumetric light-field (MesoLF) imaging of neuroactivity across cortical areas at 18 Hz
Nature Methods (2023)