Abstract
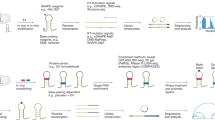
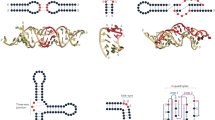
Structured noncoding RNAs underlie fundamental cellular processes, but determining their three-dimensional structures remains challenging. We demonstrate that integrating 1H NMR chemical shift data with Rosetta de novo modeling can be used to consistently determine high-resolution RNA structures. On a benchmark set of 23 noncanonical RNA motifs, including 11 'blind' targets, chemical-shift Rosetta for RNA (CS-Rosetta-RNA) recovered experimental structures with high accuracy (0.6–2.0 Å all-heavy-atom r.m.s. deviation) in 18 cases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gesteland, R.F., Cech, T. & Atkins, J.F. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA World. Vol. 43 (Cold Spring Harbor Lab Press, 2006).
Scott, L.G. & Hennig, M. Methods Mol. Biol. 452, 29–61 (2008).
Schmitz, U., James, T.L., Lukavsky, P. & Walter, P. Nat. Struct. Biol. 6, 634–638 (1999).
Jovine, L. et al. Structure 8, 527–540 (2000).
Nabuurs, S.B., Spronk, C.A.E.M., Vuister, G.W. & Vriend, G. PLoS Comput. Biol. 2, e9 (2006).
Tolbert, B.S. et al. J. Biomol. NMR 47, 205–219 (2010).
Cornilescu, G., Delaglio, F. & Bax, A. J. Biomol. NMR 13, 289–302 (1999).
Clore, G.M. & Gronenborn, A.M. Proc. Natl. Acad. Sci. USA 95, 5891–5898 (1998).
Cavalli, A., Salvatella, X., Dobson, C.M. & Vendruscolo, M. Proc. Natl. Acad. Sci. USA 104, 9615–9620 (2007).
Shen, Y. et al. Proc. Natl. Acad. Sci. USA 105, 4685–4690 (2008).
Case, D.A. J. Biomol. NMR 6, 341–346 (1995).
Cromsigt, J.A., Hilbers, C.W. & Wijmenga, S.S. J. Biomol. NMR 21, 11–29 (2001).
Girard, F.C., Ottink, O.M., Ampt, K.A., Tessari, M. & Wijmenga, S.S. Nucleic Acids Res. 35, 2800–2811 (2007).
van der Werf, R.M., Tessari, M. & Wijmenga, S.S. J. Biomol. NMR 56, 95–112 (2013).
Frank, A.T., Horowitz, S., Andricioaei, I. & Al-Hashimi, H.M. J. Phys. Chem. B 117, 2045–2052 (2013).
Das, R., Karanicolas, J. & Baker, D. Nat. Methods 7, 291–294 (2010).
Sripakdeevong, P., Kladwang, W. & Das, R. Proc. Natl. Acad. Sci. USA 108, 20573–20578 (2011).
Fleishman, S.J. & Baker, D. Cell 149, 262–273 (2012).
Deng, J., Xiong, Y., Pan, B. & Sundaralingam, M. Acta Crystallogr. D Biol. Crystallogr. 59, 1004–1011 (2003).
Leontis, N.B. & Westhof, E. RNA 7, 499–512 (2001).
Burkard, M.E. & Turner, D.H. Biochemistry 39, 11748–11762 (2000).
Nozinovic, S., Furtig, B., Jonker, H.R., Richter, C. & Schwalbe, H. Nucleic Acids Res. 38, 683–694 (2010).
Ennifar, E. et al. J. Mol. Biol. 304, 35–42 (2000).
Wu, M. & Turner, D.H. Biochemistry 35, 9677–9689 (1996).
Shankar, N. et al. Biochemistry 46, 12665–12678 (2007).
Carter, A.P. et al. Nature 407, 340–348 (2000).
Zhang, H., Fountain, M.A. & Krugh, T.R. Biochemistry 40, 9879–9886 (2001).
Aboul-ela, F., Karn, J. & Varani, G. Nucleic Acids Res. 24, 3974–3981 (1996).
Schweisguth, D.C. & Moore, P.B. J. Mol. Biol. 267, 505–519 (1997).
Lerman, Y.V. et al. RNA 17, 1664–1677 (2011).
Zhao, Q., Han, Q., Kissinger, C.R., Hermann, T. & Thompson, P.A. Acta Crystallogr. D Biol. Crystallogr. 64, 436–443 (2008).
Lukavsky, P.J., Kim, I., Otto, G.A. & Puglisi, J.D. Nat. Struct. Biol. 10, 1033–1038 (2003).
Ye, J.D. et al. Proc. Natl. Acad. Sci. USA 105, 82–87 (2008).
Davis, J.H. et al. J. Mol. Biol. 351, 371–382 (2005).
Donghi, D., Pechlaner, M., Finazzo, C., Knobloch, B. & Sigel, R.K. Nucleic Acids Res. 41, 2489–2504 (2013).
Zhao, Q. et al. Biopolymers 97, 617–628 (2012).
Ziegeler, M., Cevec, M., Richter, C. & Schwalbe, H. ChemBioChem 13, 2100–2112 (2012).
Chang, A.T. & Nikonowicz, E.P. Biochemistry 51, 3662–3674 (2012).
Kennedy, S.D., Kierzek, R. & Turner, D.H. Biochemistry 51, 9257–9259 (2012).
Acknowledgements
We thank H. Al-Hashimi for suggesting the problem and the Rosetta community for sharing code, and G. Varani and J.D. Puglisi for providing experimental 1H chemical shift data (for HIV-1 TAR apical loop and hepatitis C virus IRES subdomain IIa, respectively). Calculations were carried out on the BioX2 cluster (US National Science Foundation award CNS-0619926) and Extreme Science and Engineering Discovery Environment (XSEDE) (allocation MCB090153). We acknowledge financial support from a Burroughs Wellcome Career Award at the Scientific Interface (to R.D.), US National Institutes of Health (NIH) grant R21 GM102716 (to R.D.), a C.V. Starr Asia/Pacific Stanford Graduate Fellowship (to P.S.), NIH grant GM73969 (to E.P.N.), NIH grant GM22939 (to D.H.T.), the Swiss National Science Foundation and the European Commission (BIO-NMR, ERC-Starting Grant, Marie Curie Fellowship; to M.C.E. and R.K.O.S.), and the Deutsche Forschungsgemeinschaft (Cluster of Excellence) and the European Commission (Bio-NMR, WeNMR, Marie Curie Fellowship; to M.C., M.Z. and H.S.).
Author information
Authors and Affiliations
Contributions
P.S. and R.D. designed the research. P.S. implemented the method, generated the data, analyzed the results, and wrote the paper. R.D. assisted in analyzing the data and writing the paper. M.C., A.T.C., M.C.E., M.Z., Q.Z., G.E.F., X.G., S.D.K., R.K., E.P.N., H.S., R.K.O.S. and D.H.T. provided NMR spectroscopy data for the 11 blind targets and participated in evaluating the blinded trials. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–3, Supplementary Results and Supplementary Note (PDF 25475 kb)
Supplementary Data
Data files for the top five CS-Rosetta-RNA models, the reference experimental structures, and the experimental 1H chemical shift data for each of the 23 RNA motifs in the benchmark (included readme file provides details about the contents). (ZIP 929 kb)
Source data
Rights and permissions
About this article
Cite this article
Sripakdeevong, P., Cevec, M., Chang, A. et al. Structure determination of noncanonical RNA motifs guided by 1H NMR chemical shifts. Nat Methods 11, 413–416 (2014). https://doi.org/10.1038/nmeth.2876
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2876
This article is cited by
-
Chemical shift prediction of RNA imino groups: application toward characterizing RNA excited states
Nature Communications (2021)
-
Macromolecular modeling and design in Rosetta: recent methods and frameworks
Nature Methods (2020)
-
RNA structure refinement using NMR solvent accessibility data
Scientific Reports (2017)
-
Exploring RNA conformational space under sparse distance restraints
Scientific Reports (2017)
-
Integrative, dynamic structural biology at atomic resolution—it's about time
Nature Methods (2015)