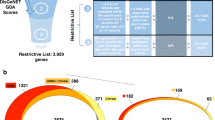
Abstract
The Drug-Gene Interaction database (DGIdb) mines existing resources that generate hypotheses about how mutated genes might be targeted therapeutically or prioritized for drug development. It provides an interface for searching lists of genes against a compendium of drug-gene interactions and potentially 'druggable' genes. DGIdb can be accessed at http://dgidb.org/.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Hopkins, A.L. & Groom, C.R. Nat. Rev. Drug Discov. 1, 727–730 (2002).
Russ, A.P. & Lampel, S. Drug Discov. Today 10, 1607–1610 (2005).
Flicek, P. et al. Nucleic Acids Res. 39, D800–D806 (2011).
Maglott, D., Ostell, J., Pruitt, K.D. & Tatusova, T. Nucleic Acids Res. 39, D52–D57 (2011).
Wang, Y. et al. Nucleic Acids Res. 40, D400–D412 (2012).
Knox, C. et al. Nucleic Acids Res. 39, D1035–D1041 (2011).
McDonagh, E.M., Whirl-Carrillo, M., Garten, Y., Altman, R.B. & Klein, T.E. Biomarkers Med. 5, 795–806 (2011).
Somaiah, N. & Simon, G.R. J. Thorac. Oncol. 6, S1758–S1785 (2011).
Rask-Andersen, M., Almen, M.S. & Schioth, H.B. Nat. Rev. Drug Discov. 10, 579–590 (2011).
Zhu, F. et al. Nucleic Acids Res. 38, D787–D791 (2010).
Ashburner, M. et al. Nat. Genet. 25, 25–29 (2000).
Kumar, R.D., Chang, L.W., Ellis, M.J. & Bose, R. PLoS One 8, e67980 (2013).
Banerji, S. et al. Nature 486, 405–409 (2012).
Cancer Genome Atlas Network. Nature 490, 61–70 (2012).
Kan, Z. et al. Nature 466, 869–873 (2010).
Shah, S.P. et al. Nature 486, 395–399 (2012).
Stephens, P.J. et al. Nature 486, 400–404 (2012).
Bose, R. et al. Cancer Discov. 3, 224–237 (2013).
Yeh, P. et al. Clin. Cancer Res. 19, 1894–1901 (2013).
Hunter, S. et al. Nucleic Acids Res. 40, D306–D312 (2012).
Punta, M. et al. Nucleic Acids Res. 40, D290–D301 (2012).
Kuhn, M. et al. Nucleic Acids Res. 40, D876–D880 (2012).
Hecker, N. et al. Nucleic Acids Res. 40, D1113–D1117 (2012).
Gaulton, A. et al. Nucleic Acids Res. 40, D1100–D1107 (2012).
von Eichborn, J. et al. Nucleic Acids Res. 39, D1060–D1066 (2011).
Davis, A.P. et al. Nucleic Acids Res. 41, D1104–D1114 (2013).
Gao, Z. et al. BMC Bioinformatics 9, 104 (2008).
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanam, S. Science 298, 1912–1934 (2002).
Orth, A.P., Batalov, S., Perrone, M. & Chanda, S.K. Expert Opin. Ther. Targets 8, 587–596 (2004).
Lamb, J. et al. Science 313, 1929–1935 (2006).
Liu, T., Lin, Y., Wen, X., Jorissen, R.N. & Gilson, M.K. Nucleic Acids Res. 35, D198–D201 (2007).
Yang, W. et al. Nucleic Acids Res. 41, D955–D961 (2013).
Barretina, J. et al. Nature 483, 603–607 (2012).
Lim, E. et al. Nucleic Acids Res. 38, D781–D786 (2010).
Preissner, S. et al. Nucleic Acids Res. 38, D237–D243 (2010).
Kuhn, M., Campillos, M., Letunic, I., Jensen, L.J. & Bork, P. Mol. Syst. Biol. 6, 343 (2010).
Lounkine, E. et al. Nature 486, 361–367 (2012).
Acknowledgements
We thank the creators of each source we used as input and all users of DGIdb who have contributed feedback, R. Altman, Stanford University and PharmGKB for granting us a license to mine their data, W. Pao of My Cancer Genome for agreeing to collaborate with us, and the individuals who participated in research that led to the breast cancer data used to demonstrate the utility of DGIdb. This work was funded by an US National Institutes of Health NHGRI center grant (U54 HG003079) to R.K.W.
Author information
Authors and Affiliations
Contributions
O.L.G. and M.G. wrote the manuscript, were responsible for the selection and manual curation of data sources, creation of importers, initial design of the web interface, testing, analysis and figure creation for the manuscript. I.D. and J.F.M. helped create figures. A.C.C. and J.V.W. conceived and lead the Ruby implementation of the web interface, database, importers and application programming interface (API). J.K., M.G., O.L.G., S.M.S., I.D. and J.F.M. made contributions to the code. J.F.M. was the lead user experience web developer. S.M.S., J.M.E., J.R.W. and D.J.D. supervised software development. M.B.C. facilitated the public deployment of DGIdb and all related systems requirements. R.B., R.D.K., C.A.M. and D.E.L. provided beta testing feedback. J.S. and R.G. provided and curated the 'targeted agents in lung cancer' data set. R.B. and R.D.K. contributed the dGene data set and the breast cancer data used for analysis demonstration purposes. N.C.S. contributed manual curation of data sources and documentation. S.M.S., J.M.E., D.J.D., L.D., T.J.L., E.R.M. and R.K.W. contributed ideas and use case requirements. A.C.C., J.V.W., C.A.M., R.D.K., R.B., D.E.L. and E.R.M. contributed text and revised the manuscript. E.R.M. and R.K.W. are mentors of M.G. and O.L.G.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–4 (PDF 7218 kb)
Supplementary Table 5
Breast cancer druggable candidates. (XLSX 51 kb)
Supplementary Table 6
Breast cancer known druggability by patient. (XLSX 1114 kb)
Rights and permissions
About this article
Cite this article
Griffith, M., Griffith, O., Coffman, A. et al. DGIdb: mining the druggable genome. Nat Methods 10, 1209–1210 (2013). https://doi.org/10.1038/nmeth.2689
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2689
This article is cited by
-
Discovery of therapeutic targets for spinal cord injury based on molecular mechanisms of axon regeneration after conditioning lesion
Journal of Translational Medicine (2023)
-
Network analyses unveil ageing-associated pathways evolutionarily conserved from fungi to animals
GeroScience (2023)
-
Enhancer promoter interactome and Mendelian randomization identify network of druggable vascular genes in coronary artery disease
Human Genomics (2022)
-
Multi-site desmoplastic small round cell tumors are genetically related and immune-cold
npj Precision Oncology (2022)
-
Analysis of multiple databases identifies crucial genes correlated with prognosis of hepatocellular carcinoma
Scientific Reports (2022)