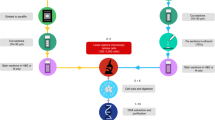
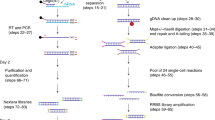
Abstract
Tissue gene expression profiling is performed on homogenates or on populations of isolated single cells to resolve molecular states of different cell types. In both approaches, histological context is lost. We have developed an in situ sequencing method for parallel targeted analysis of short RNA fragments in morphologically preserved cells and tissue. We demonstrate in situ sequencing of point mutations and multiplexed gene expression profiling in human breast cancer tissue sections.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Levsky, J.M. & Singer, R.H. Trends Cell Biol. 13, 4–6 (2003).
Wang, Z., Gerstein, M. & Snyder, M. Nat. Rev. Genet. 10, 57–63 (2009).
Bonner, R.F. et al. Science 278, 1481–1483 (1997).
Dalerba, P. et al. Nat. Biotechnol. 29, 1120–1127 (2011).
Navin, N. et al. Nature 472, 90–94 (2011).
Tang, F. et al. Nat. Methods 6, 377–382 (2009).
Hou, Y. et al. Cell 148, 873–885 (2012).
Xu, X. et al. Cell 148, 886–895 (2012).
Nilsson, M. et al. Science 265, 2085–2088 (1994).
Banér, J., Nilsson, M., Mendel-Hartvig, M. & Landegren, U. Nucleic Acids Res. 26, 5073–5078 (1998).
Larsson, C., Grundberg, I., Soderberg, O. & Nilsson, M. Nat. Methods 7, 395–397 (2010).
Shendure, J. et al. Science 309, 1728–1732 (2005).
Drmanac, R. et al. Science 327, 78–81 (2010).
Kamentsky, L. et al. Bioinformatics 27, 1179–1180 (2011).
Thévenaz, P., Ruttimann, U.E. & Unser, M. IEEE Trans. Image Process. 7, 27–41 (1998).
Sparano, J.A. & Paik, S. J. Clin. Oncol. 26, 721–728 (2008).
Wang, E.T. et al. Nature 456, 470–476 (2008).
Acknowledgements
We thank J. Lee, G.M. Church and F. Pontén for valuable discussion about this work. We thank M. Dahlberg for helping extracting RNA-seq data from publications. The research reported in this paper was funded by the Swedish Research Council, VINNOVA project “Companion diagnostic initiative,” the European Community's 7th Framework Program (FP7/2007-2013) under grant agreement nos. 259796 (DiaTools) and 201418 (READNA), the Science for Life Laboratory, Stockholm and Uppsala, and the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115234 (OncoTrack).
Author information
Authors and Affiliations
Contributions
R.K. and M.M. designed and performed the experiments. J.B. provided tissue sections and pathology examination of the tissue. C.W. designed the image analysis pipelines and performed the image analysis together with A.P. J.S. performed the correlation between in situ sequencing and RNA-seq data. R.K., M.M., C.W. and M.N. wrote the manuscript. All authors commented on and revised the manuscript. M.N. conceived the idea and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
M.N. owns shares in the company Olink AB, Uppsala, Sweden, which holds patents whose value may be affected by publication of these results.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1–7 and Supplementary Notes 1 and 2 (PDF 3726 kb)
Source data
Rights and permissions
About this article
Cite this article
Ke, R., Mignardi, M., Pacureanu, A. et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods 10, 857–860 (2013). https://doi.org/10.1038/nmeth.2563
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2563
This article is cited by
-
Next-Generation Sequencing in Medicinal Plants: Recent Progress, Opportunities, and Challenges
Journal of Plant Growth Regulation (2024)
-
STmut: a framework for visualizing somatic alterations in spatial transcriptomics data of cancer
Genome Biology (2023)
-
In silico tissue generation and power analysis for spatial omics
Nature Methods (2023)
-
Heterogeneity of the tumor immune microenvironment and clinical interventions
Frontiers of Medicine (2023)
-
Spatiotemporal Dynamics of the Molecular Expression Pattern and Intercellular Interactions in the Glial Scar Response to Spinal Cord Injury
Neuroscience Bulletin (2023)