Abstract
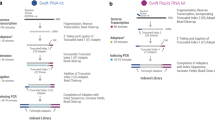
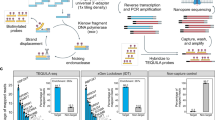
RNA-seq is an effective method for studying the transcriptome, but it can be difficult to apply to scarce or degraded RNA from fixed clinical samples, rare cell populations or cadavers. Recent studies have proposed several methods for RNA-seq of low-quality and/or low-quantity samples, but the relative merits of these methods have not been systematically analyzed. Here we compare five such methods using metrics relevant to transcriptome annotation, transcript discovery and gene expression. Using a single human RNA sample, we constructed and sequenced ten libraries with these methods and compared them against two control libraries. We found that the RNase H method performed best for chemically fragmented, low-quality RNA, and we confirmed this through analysis of actual degraded samples. RNase H can even effectively replace oligo(dT)-based methods for standard RNA-seq. SMART and NuGEN had distinct strengths for measuring low-quantity RNA. Our analysis allows biologists to select the most suitable methods and provides a benchmark for future method development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Referenced accessions
NCBI Reference Sequence
Change history
02 December 2013
In the version of this article initially published, in the Online Methods "RNase H libraries" section, the sentence beginning with "We added 5 μl preheated RNase H...." should have read "We added 5 μl preheated RNase H reaction mix that contains 10 U of Hybridase Thermostable RNase H (Epicentre), 0.5 μmol Tris-HCl, pH 7.5, 1 μmol NaCl and 0.2 μmol MgCl2 to the RNA and DNA oligo mix, incubated this mixture at 45 °C for 30 min and then placed it on ice." The errors have been corrected in the HTML and PDF versions of this article.
References
Aviv, H. & Leder, P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc. Natl. Acad. Sci. USA 69, 1408–1412 (1972).
Yang, L., Duff, M.O., Graveley, B.R., Carmichael, G.G. & Chen, L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 12, R16 (2011).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Ramsköld, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Islam, S. et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 21, 1160–1167 (2011).
Sinicropi, D. & Morlan, J. Methods for depleting RNA from nucleic acid samples. US patent application 20110111409 (2011).
Morlan, J.D., Qu, K. & Sinicropi, D.V. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS ONE 7, e42882 (2012).
Huang, R. et al. An RNA-Seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PLoS ONE 6, e27288 (2011).
Yi, H. et al. Duplex-specific nuclease efficiently removes rRNA for prokaryotic RNA-seq. Nucleic Acids Res. 39, e140 (2011).
Tariq, M.A., Kim, H.J., Jejelowo, O. & Pourmand, N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 39, e120 (2011).
Levin, J.Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 (2010).
DeLuca, D.S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Beyer, A.L. & Osheim, Y.N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2, 754–765 (1988).
Yang, Y.H. et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15 (2002).
Aird, D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12, R18 (2011).
Rosenkranz, R., Borodina, T., Lehrach, H. & Himmelbauer, H. Characterizing the mouse ES cell transcriptome with Illumina sequencing. Genomics 92, 187–194 (2008).
Giannoukos, G. et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 13, R23 (2012).
Griffin, M., Abu-El-Haija, M., Abu-El-Haija, M., Rokhlina, T. & Uc, A. Simplified and versatile method for isolation of high-quality RNA from pancreas. Biotechniques 52, 332–334 (2012).
Pan, X. et al. Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc. Natl. Acad. Sci. USA 110, 594–599 (2013).
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J.L. & Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Maden, B.E. et al. Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem. J. 246, 519–527 (1987).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Dreszer, T.R. et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 40, D918–D923 (2012).
Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Use R!) (Springer, New York, 2009).
Acknowledgements
We thank D. Sinicropi for alerting us to his RNase H method, Broad Genomics Platform for sequencing work, C. Nusbaum, R. Johnson, M. Yassour and V. Savova for helpful discussions, and L. Gaffney for assistance in drafting figures. Work was supported by US National Institutes of Health Pioneer Award DP1-OD003958-01, US National Human Genome Research Institute (NHGRI) 1P01HG005062-01, NHGRI Center of Excellence in Genome Science Award 1P50HG006193-01, the Howard Hughes Medical Institute, the Merkin Foundation for Stem Cell Research and the Klarman Cell Observatory at the Broad Institute (to A.R.), and by NHGRI grant HG03067. N.P. was supported by a postdoctoral research fellowship of the Fund for Scientific Research–Flanders (FWO Vlaanderen).
Author information
Authors and Affiliations
Contributions
J.Z.L., X.A. and A.R. conceived the research. X.A. prepared the cDNA libraries. D.B.-R., R.S., N.P., M.A.B. and A.R. developed and performed computational analysis. D.S.D. contributed code. D.S.D., A.M.B., A.S., A.W. and T.F. helped with computational analysis. D.A.T., N.P., A.R. and J.Z.L. supervised the research. J.Z.L., X.A., D.B.-R. and A.R. wrote the paper. R.S., A.G. and D.S.D. assisted in editing the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures (i)
Supplementary Figures 1–7 (PDF 45665 kb)
Supplementary Text and Figures (ii)
Supplementary Figures 8 and 9 (PDF 20201 kb)
Supplementary Text and Figures (iii)
Supplementary Tables 1–5 and 7 and Supplementary Notes 1–5 (PDF 1010 kb)
Supplementary Table 6
RNase H oligonucleotide sequences (XLSX 19 kb)
Rights and permissions
About this article
Cite this article
Adiconis, X., Borges-Rivera, D., Satija, R. et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods 10, 623–629 (2013). https://doi.org/10.1038/nmeth.2483
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2483
This article is cited by
-
Distinct forebrain regions define a dichotomous astrocytic profile in multiple system atrophy
Acta Neuropathologica Communications (2024)
-
RIP-PEN-seq identifies a class of kink-turn RNAs as splicing regulators
Nature Biotechnology (2024)
-
T-RHEX-RNAseq – a tagmentation-based, rRNA blocked, random hexamer primed RNAseq method for generating stranded RNAseq libraries directly from very low numbers of lysed cells
BMC Genomics (2023)
-
A comprehensive assessment of exome capture methods for RNA sequencing of formalin-fixed and paraffin-embedded samples
BMC Genomics (2023)
-
Optimizing identification of consensus molecular subtypes in muscle-invasive bladder cancer: a comparison of two sequencing methods and gene sets using FFPE specimens
BMC Cancer (2023)