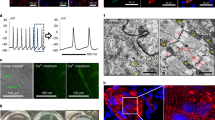
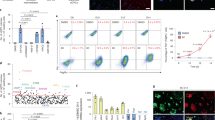
Abstract
Lineage conversion of one somatic cell type to another is an attractive approach for generating specific human cell types. Lineage conversion can be direct, in the absence of proliferation and multipotent progenitor generation, or indirect, by the generation of expandable multipotent progenitor states. We report the development of a reprogramming methodology in which cells transition through a plastic intermediate state, induced by brief exposure to reprogramming factors, followed by differentiation. We use this approach to convert human fibroblasts to mesodermal progenitor cells, including by non-integrative approaches. These progenitor cells demonstrated bipotent differentiation potential and could generate endothelial and smooth muscle lineages. Differentiated endothelial cells exhibited neo-angiogenesis and anastomosis in vivo. This methodology for indirect lineage conversion to angioblast-like cells adds to the armamentarium of reprogramming approaches aimed at the study and treatment of ischemic pathologies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Change history
07 February 2020
"An amendment to this paper has been published and can be accessed via a link at the top of the paper."
References
Sancho-Martinez, I., Nivet, E. & Izpisua Belmonte, J.C. The labyrinth of nuclear reprogramming. J. Mol. Cell Biol. 3, 327–329 (2011).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Laiosa, C.V., Stadtfeld, M., Xie, H., de Andres-Aguayo, L. & Graf, T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25, 731–744 (2006).
Ieda, M. et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 (2010).
Vierbuchen, T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010).
Vierbuchen, T. & Wernig, M. Direct lineage conversions: unnatural but useful? Nat. Biotechnol. 29, 892–907 (2011).
Efe, J.A. et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215–222 (2011).
Kim, J. et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA 108, 7838–7843 (2011).
Sancho-Martinez, I., Baek, S.H. & Izpisua Belmonte, J.C. Lineage conversion methodologies meet the reprogramming toolbox. Nat. Cell Biol. 14, 892–899 (2012).
Papp, B. & Plath, K. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res. 21, 486–501 (2011).
Plath, K. & Lowry, W.E. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 12, 253–265 (2011).
Goldman, O. et al. A boost of BMP4 accelerates the commitment of human embryonic stem cells to the endothelial lineage. Stem Cells 27, 1750–1759 (2009).
Dzierzak, E. & Speck, N.A. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129–136 (2008).
Choi, K.D. et al. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells 27, 559–567 (2009).
James, D. et al. Expansion and maintenance of human embryonic stem cell–derived endothelial cells by TGFβ inhibition is Id1 dependent. Nat. Biotechnol. 28, 161–166 (2010).
Levenberg, S., Ferreira, L.S., Chen-Konak, L., Kraehenbuehl, T.P. & Langer, R. Isolation, differentiation and characterization of vascular cells derived from human embryonic stem cells. Nat. Protoc. 5, 1115–1126 (2010).
Anokye-Danso, F. et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376–388 (2011).
Subramanyam, D. et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 29, 443–448 (2011).
Narazaki, G. et al. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation 118, 498–506 (2008).
Cheung, C., Bernardo, A.S., Trotter, M.W., Pedersen, R.A. & Sinha, S. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat. Biotechnol. 30, 165–173 (2012).
Okita, K. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 (2011).
Chan, E.M. et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 27, 1033–1037 (2009).
Ohta, T., Ito, S. & Nakazato, Y. All-or-nothing responses to carbachol in single intestinal smooth muscle cells of rat. Br. J. Pharmacol. 112, 972–976 (1994).
Kohda, M., Komori, S., Unno, T. & Ohashi, H. Carbachol-induced [Ca2+]i oscillations in single smooth muscle cells of guinea-pig ileum. J. Physiol. (Lond.) 492, 315–328 (1996).
Yamanaka, S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10, 678–684 (2012).
Yusa, K. et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature 478, 391–394 (2011).
Ginsberg, M. et al. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFβ suppression. Cell 151, 559–575 (2012).
Giorgetti, A. et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell 5, 353–357 (2009).
Aasen, T. et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276–1284 (2008).
Ludwig, T.E. et al. Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 24, 185–187 (2006).
Acknowledgements
We are thankful to Y. Zheng for his expertise and assistance with sorting procedures, C. Maiza for his expertise and assistance with in vivo procedures and M. Schwarz for administrative support. L.K. was partially supported by the California Institute for Regenerative Medicine. E.N. was partially supported by an F.M. Kirby Foundation postdoctoral fellowship. A.M.D. was supported by the Helmsley Foundation. L.C.L., R.D.T., F.S.B. and J.F.L. are supported by the California Institute for Regenerative Medicine (CL1-00502, RT1-01108, TR1-01250, RN2-00931), US National Institutes of Health (R33MH087925), US National Institutes of Health/National Institute Child Health and Human Development K12 Career Development Award (L.C.L.), Hartwell Foundation (L.C.L., R.D.T., F.S.B.), Millipore Foundation (J.F.L.) and Esther O'Keefe Foundation (J.F.L.). Work in the laboratory of F.H.G. was supported by the JPB Foundation, G. Harold and Leila Y. Mathers Charitable Foundation and Ellison Medical Foundation. Work in the laboratory of J.C.I.B. was supported by grants from Fundacion Cellex, the G. Harold and Leila Y. Mathers Charitable Foundation, the Leona M. and Harry B. Helmsley Charitable Trust, Sanofi, Ministerio de Economia y Competitividad (PLE2009-0100), Instituto de Salud Carlos III (ISCIII), Terapia Celular (TerCel) (RD06/0010/0016) and Fondo Europeo de Desarrollo Regional (FEDER).
Author information
Authors and Affiliations
Contributions
L.K., I.S.-M., E.N. and J.C.I.B. designed all experiments. I.S.-M., E.N. and J.C.I.B. wrote the manuscript. L.K., I.S.-M., K.M., E.N. and A.A. performed and analyzed all experiments. C.P., C.V.-C., F.B., E.N., I.D. and J.-M.H. performed in vivo experiments. K.M. was responsible for all cell culture–related work. M.L., A.M.D. and F.H.G. provided microRNA constructs and reagents. J.P., Y.X., S.R., I.D., N.M., C.R., A.M.D. and F.H.G. contributed to the overall design of the project. F.S.B., R.D.T., J.F.L. and L.C.L. performed and analyzed genome-wide array DNA methylation and gene expression studies.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 and Supplementary Tables 1 and 3 (PDF 23185 kb)
Supplementary Table 2
Specific mRNA fold change of the qPCR data summarized in Figures 1–3. (XLSX 54 kb)
Supplementary Data
Detailed mRNA and methylation array data including differentially regulated gene lists. (XLSX 22235 kb)
Rights and permissions
About this article
Cite this article
Kurian, L., Sancho-Martinez, I., Nivet, E. et al. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat Methods 10, 77–83 (2013). https://doi.org/10.1038/nmeth.2255
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2255
This article is cited by
-
Mechanisms, pathways and strategies for rejuvenation through epigenetic reprogramming
Nature Aging (2023)
-
Tumor bud-derived CCL5 recruits fibroblasts and promotes colorectal cancer progression via CCR5-SLC25A24 signaling
Journal of Experimental & Clinical Cancer Research (2022)
-
Genome-wide CRISPR/Cas9 screening in human iPS derived cardiomyocytes uncovers novel mediators of doxorubicin cardiotoxicity
Scientific Reports (2021)
-
Direct conversion of human fibroblasts into therapeutically active vascular wall-typical mesenchymal stem cells
Cellular and Molecular Life Sciences (2020)
-
Conversion of human adipose-derived stem cells into functional and expandable endothelial-like cells for cell-based therapies
Stem Cell Research & Therapy (2018)