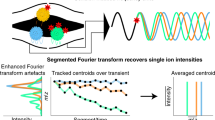
Abstract
The analysis of intact protein assemblies in native-like states by mass spectrometry offers a wealth of information on their biochemical and biophysical properties. Here we show that the Orbitrap mass analyzer can be used to measure protein assemblies of molecular weights approaching one megadalton with sensitivity down to the detection of single ions. Minor instrumental modifications enabled the measurement of various protein assemblies with outstanding mass-spectral resolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sharon, M. & Robinson, C.V. Annu. Rev. Biochem. 76, 167–193 (2007).
Heck, A.J. Nat. Methods 5, 927–933 (2008).
Shoemaker, G.K. et al. Mol. Cell. Proteomics 9, 1742–1751 (2010).
Sobott, F., Hernandez, H., McCammon, M.G., Tito, M.A. & Robinson, C.V. Anal. Chem. 74, 1402–1407 (2002).
van den Heuvel, R.H. et al. Anal. Chem. 78, 7473–7483 (2006).
Christ, P. et al. Eur. J. Mass Spectrom. (Chichester, Eng.) 10, 469–476 (2004).
Chernushevich, I.V. & Thomson, B.A. Anal. Chem. 76, 1754–1760 (2004).
Zhang, Z., Pan, H. & Chen, X. Mass Spectrom. Rev. 28, 147–176 (2009).
Perry, R.H., Cooks, R.G. & Noll, R.J. Mass Spectrom. Rev. 27, 661–699 (2008).
Hendrix, R.W. & Johnson, J.E. Adv. Exp. Med. Biol. 726, 351–363 (2012).
Bochtler, M., Ditzel, L., Groll, M., Hartmann, C. & Huber, R. Annu. Rev. Biophys. Biomol. Struct. 28, 295–317 (1999).
Krishna, K.A., Rao, G.V. & Rao, K.R. Curr. Protein Pept. Sci. 8, 418–425 (2007).
Rostom, A.A. et al. Proc. Natl. Acad. Sci. USA 97, 5185–5190 (2000).
Tito, M.A., Tars, K., Valegard, K., Hadju, J. & Robinson, C.V. J. Am. Chem. Soc. 122, 3550–3551 (2000).
Makarov, A. & Denisov, E. J. Am. Soc. Mass Spectrom. 20, 1486–1495 (2009).
Benesch, J.L., Aquilina, J.A., Ruotolo, B.T., Sobott, F. & Robinson, C.V. Chem. Biol. 13, 597–605 (2006).
Monti, M.C. et al. Nucleic Acids Res. 39, 8052–8064 (2011).
Rogniaux, H. et al. Anal. Biochem. 291, 48–61 (2001).
Zhou, M. et al. Science 334, 380–385 (2011).
Cui, W., Rohrs, H.W. & Gross, M.L. Analyst (Lond.) 136, 3854–3864 (2011).
Quaite-Randall, E. & Joachimiak, A. Methods Mol. Biol. 140, 29–39 (2000).
Olsen, J.V. et al. Mol. Cell. Proteomics 8, 2759–2769 (2009).
Michalski, A. et al. Mol. Cell. Proteomics 10, M111.011015 (2011).
Makarov, A. Anal. Chem. 72, 1156–1162 (2000).
Acknowledgements
This work was supported in part by the PRIME-XS project, Grant Agreement Number 262067, and by the PROSPECTS network (grant HEALTH-F4-2008-201648) both funded by the European Union Seventh Framework Program. The Netherlands Proteomics Centre, embedded in The Netherlands Genomics Initiative, is acknowledged for funding. The authors thank Michael Gröll (Technische Universitat Munchen) for his kind donation of the 20S proteasome sample, Jack Johnson (Scripps Research Institute) for the HK97 capsomer sample, Genmab for the IgG antibody, and Uwe Rickens (Thermo Fisher Scientific) for developing IonDetectionConverter software for the single ion experiments.
Author information
Authors and Affiliations
Contributions
R.J.R., E.Da., E.De., A.M. and A.J.R.H. performed the experiments and wrote the manuscript. A.M. supervised the modifications on the Orbitrap mass analyzer. A.M. and A.J.R.H. conceptually designed the work.
Corresponding author
Ethics declarations
Competing interests
E.Da., E.De. and A.M. are employees of Thermo Fisher Scientific, the manufacturer of the Exactive Plus instrument used in this research.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 950 kb)
Rights and permissions
About this article
Cite this article
Rose, R., Damoc, E., Denisov, E. et al. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods 9, 1084–1086 (2012). https://doi.org/10.1038/nmeth.2208
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2208
This article is cited by
-
Analysis of Fatty Acid in Biological Samples Using Liquid Chromatography–Quadrupole-Orbitrap Mass Spectrometry Under Parallel Reaction Monitoring Mode
Journal of Analysis and Testing (2023)
-
Three-dimensional structure determination of protein complexes using matrix-landing mass spectrometry
Nature Communications (2022)
-
Frequency chasing of individual megadalton ions in an Orbitrap analyser improves precision of analysis in single-molecule mass spectrometry
Nature Chemistry (2022)
-
Studying protein structure and function by native separation–mass spectrometry
Nature Reviews Chemistry (2022)
-
The emerging landscape of single-molecule protein sequencing technologies
Nature Methods (2021)