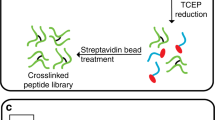
Abstract
The mass spectrometric identification of chemically cross-linked peptides (CXMS) specifies spatial restraints of protein complexes; these values complement data obtained from common structure-determination techniques. Generic methods for determining false discovery rates of cross-linked peptide assignments are currently lacking, thus making data sets from CXMS studies inherently incomparable. Here we describe an automated target-decoy strategy and the software tool xProphet, which solve this problem for large multicomponent protein complexes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Young, M.M. et al. Proc. Natl. Acad. Sci. USA 97, 5802–5806 (2000).
Rappsilber, J. et al. Anal. Chem. 72, 267–275 (2000).
Leitner, A. et al. Mol. Cell. Proteomics 9, 1634–1649 (2010).
Rappsilber, J. J. Struct. Biol. 173, 530–540 (2011).
Sinz, A. Mass Spectrom. Rev. 25, 663–682 (2006).
Back, J.W. et al. J. Mol. Biol. 331, 303–313 (2003).
Mayne, S.L. & Patterton, H.G. Brief. Bioinform. 12, 660–671 (2011).
Lasker, K. et al. Proc. Natl. Acad. Sci. USA 109, 1380–1387 (2012).
Chen, Z.A. et al. EMBO J. 29, 717–726 (2010).
Bohn, S. et al. Proc. Natl. Acad. Sci. USA 107, 20992–20997 (2010).
Jennebach, S. et al. Nucleic Acids Res. doi:10.1093/nar/gks220 (6 March 2012).
Rinner, O. et al. Nat. Methods 5, 315–318 (2008).
Elias, J.E. & Gygi, S.P. Nat. Methods 4, 207–214 (2007).
Leitner, A. et al. Mol. Cell Proteomics 11, M111.014126 (2012).
Petrotchenko, E.V. & Borchers, C.H. Mass Spectrom. Rev. 29, 862–876 (2010).
Pettersen, E.F. et al. J. Comput. Chem. 25, 1605–1612 (2004).
Pieper, U. et al. Nucleic Acids Res. 30, 255–259 (2002).
Efron, B. & Tibshirani, R. Genet. Epidemiol. 23, 70–86 (2002).
Acknowledgements
This work was supported by the European Union 7th Framework project PROSPECTS (Proteomics Specification in Space and Time, grant HEALTH-F4-2008-201648) and ERC advanced grant 'Proteomics v3.0' (grant no. 233226) of the European Union to R.A. We thank O. Rinner and A.I. Nesvizhskii for constructive discussions.
Author information
Authors and Affiliations
Contributions
T.W. and A.L. performed cross-linking experiments and mass spectrometry analysis. T.W. and M.C. developed the target-decoy algorithm. T.W. analyzed the data and designed and wrote the software. S.B. purified 26S proteasomes; F.F. provided the structural model of the 26S proteasome; and T.W., A.L., S.B., M.C., F.H., F.F., M.B. and R.A. discussed and designed experiments. All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1, Supplementary Results 1–3 and Supplementary Methods (PDF 522 kb)
Supplementary Table 2
Identifications of the 8-mix data set. (XLSX 49 kb)
Supplementary Table 3
Identifications of the 26S data set. (XLSX 47 kb)
Rights and permissions
About this article
Cite this article
Walzthoeni, T., Claassen, M., Leitner, A. et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods 9, 901–903 (2012). https://doi.org/10.1038/nmeth.2103
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2103